Di-valent siRNA-mediated silencing of MSH3 blocks somatic repeat expansion in mouse models of Huntington's disease
- PMID: 37177784
- PMCID: PMC10277892
- DOI: 10.1016/j.ymthe.2023.05.006
Di-valent siRNA-mediated silencing of MSH3 blocks somatic repeat expansion in mouse models of Huntington's disease
Erratum in
-
Di-valent siRNA-mediated silencing of MSH3 blocks somatic repeat expansion in mouse models of Huntington's disease.Mol Ther. 2023 Nov 1;31(11):3355-3356. doi: 10.1016/j.ymthe.2023.09.016. Epub 2023 Sep 25. Mol Ther. 2023. PMID: 37751745 Free PMC article. No abstract available.
Abstract
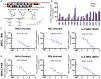
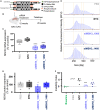
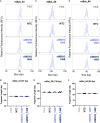
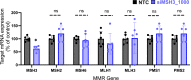
Huntington's disease (HD) is a severe neurodegenerative disorder caused by the expansion of the CAG trinucleotide repeat tract in the huntingtin gene. Inheritance of expanded CAG repeats is needed for HD manifestation, but further somatic expansion of the repeat tract in non-dividing cells, particularly striatal neurons, hastens disease onset. Called somatic repeat expansion, this process is mediated by the mismatch repair (MMR) pathway. Among MMR components identified as modifiers of HD onset, MutS homolog 3 (MSH3) has emerged as a potentially safe and effective target for therapeutic intervention. Here, we identify a fully chemically modified short interfering RNA (siRNA) that robustly silences Msh3 in vitro and in vivo. When synthesized in a di-valent scaffold, siRNA-mediated silencing of Msh3 effectively blocked CAG-repeat expansion in the striatum of two HD mouse models without affecting tumor-associated microsatellite instability or mRNA expression of other MMR genes. Our findings establish a promising treatment approach for patients with HD and other repeat expansion diseases.
Keywords: CAG expansion disorders; DNA instability; Huntington’s disease; mismatch repair; neurodegeneration; oligonucleotide therapeutics; siRNA.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.K. and N.A. are co-founders of Atalanta. A.K. and N.A. are on the scientific advisory board of Atalanta. N.A. is on the scientific advisory board of Huntington’s Disease Society for America and that of Biogen and has licensing to Spark Therapeutics. A.K. has a financial interest in Atalanta. The authors have patents on MSH3 as a therapeutic target and the methodology described in the paper.
Figures


Comment in
-
Can MSH3 lowering stop HTT repeat expansion in its CAG tract?Mol Ther. 2023 Jun 7;31(6):1509-1511. doi: 10.1016/j.ymthe.2023.05.010. Epub 2023 May 25. Mol Ther. 2023. PMID: 37236185 Free PMC article. No abstract available.
References
-
- Tabrizi S.J., Estevez-Fraga C., van Roon-Mom W.M.C., Flower M.D., Scahill R.I., Wild E.J., Muñoz-Sanjuan I., Sampaio C., Rosser A.E., Leavitt B.R. Potential disease-modifying therapies for Huntington's disease: lessons learned and future opportunities. Lancet Neurol. 2022;21:645–658. doi: 10.1016/S1474-4422(22)00121-1. - DOI - PMC - PubMed
-
- Ghosh R., Tabrizi S.J. In: Polyglutamine Disorders. Nóbrega C., Pereira de Almeida L., editors. Springer International Publishing; 2018. Clinical features of huntington’s disease; pp. 1–28. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

