Identification of structurally diverse FSP1 inhibitors that sensitize cancer cells to ferroptosis
- PMID: 37178691
- PMCID: PMC10524360
- DOI: 10.1016/j.chembiol.2023.04.007
Identification of structurally diverse FSP1 inhibitors that sensitize cancer cells to ferroptosis
Abstract
Ferroptosis is a regulated form of cell death associated with the iron-dependent accumulation of phospholipid hydroperoxides. Inducing ferroptosis is a promising approach to treat therapy-resistant cancer. Ferroptosis suppressor protein 1 (FSP1) promotes ferroptosis resistance in cancer by generating the antioxidant form of coenzyme Q10 (CoQ). Despite the important role of FSP1, few molecular tools exist that target the CoQ-FSP1 pathway. Through a series of chemical screens, we identify several structurally diverse FSP1 inhibitors. The most potent of these compounds, ferroptosis sensitizer 1 (FSEN1), is an uncompetitive inhibitor that acts selectively through on-target inhibition of FSP1 to sensitize cancer cells to ferroptosis. Furthermore, a synthetic lethality screen reveals that FSEN1 synergizes with endoperoxide-containing ferroptosis inducers, including dihydroartemisinin, to trigger ferroptosis. These results provide new tools that catalyze the exploration of FSP1 as a therapeutic target and highlight the value of combinatorial therapeutic regimes targeting FSP1 and additional ferroptosis defense pathways.
Keywords: FSP1; GPX4; cancer; cell death; coenzyme Q10; endoperoxide; ferroptosis; glutathione; lipid peroxidation; small molecule screen.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests J.A.O. is a member of the scientific advisory board for Vicinitas Therapeutics. S.J.D. is a member of the scientific advisory board for Ferro Therapeutics and Hillstream BioPharma, Inc. J.S. is a member of the board of directors for Zenith Therapeutics and a scientific advisor to Lyterian Biosciences and Organos. S.J.D., J.A.O., J.M.H., E.W., J.S., C.E.D., and K.B. have ferroptosis-related patent applications.
Figures
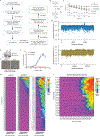
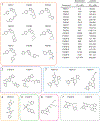

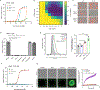
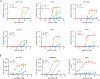


Comment in
-
Sabotaging the breaks: FSEN1 expands the toolbox of FSP1 inhibitors.Cell Chem Biol. 2023 Sep 21;30(9):1006-1008. doi: 10.1016/j.chembiol.2023.08.015. Cell Chem Biol. 2023. PMID: 37738951
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

