Genome-wide analysis of RNA-binding proteins co-expression with alternative splicing events in mitral valve prolapse
- PMID: 37180137
- PMCID: PMC10171460
- DOI: 10.3389/fimmu.2023.1078266
Genome-wide analysis of RNA-binding proteins co-expression with alternative splicing events in mitral valve prolapse
Abstract
Objectives: We investigated the role and molecular mechanisms of RNA-binding proteins (RBPs) and their regulated alternative splicing events (RASEs) in the pathogenesis of mitral valve prolapse (MVP).
Methods: For RNA extraction, we obtained peripheral blood mononuclear cells (PBMCs) from five patients with MVP, with or without chordae tendineae rupture, and five healthy individuals. High-throughput sequencing was used for RNA sequencing (RNA-seq). Differentially expressed genes (DEGs) analysis, alternative splicing (AS) analysis, functional enrichment analysis, co-expression of RBPs, and alternative splicing events (ASEs) analysis were conducted.
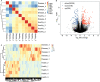
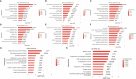
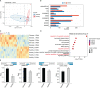
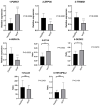
Results: The MVP patients exhibited 306 up-regulated genes and 198 down-regulated genes. All down- and up-regulated genes were enriched in both Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Furthermore, MVP was closely associated with the top 10 enriched terms and pathways. In MVP patients, 2,288 RASEs were found to be significantly different, and four suitable RASEs (CARD11 A3ss, RBM5 ES, NCF1 A5SS, and DAXX A3ss) were tested. We identified 13 RNA-binding proteins (RBPs) from the DEGs and screened out four RBPs (ZFP36, HSPA1A, TRIM21, and P2RX7). We selected four RASEs based on the co-expression analyses of RBPs and RASEs, including exon skipping (ES) of DEDD2, alternative 3' splice site (A3SS) of ETV6, mutually exclusive 3'UTRs (3pMXE) of TNFAIP8L2, and A3SS of HLA-B. Furthermore, the selected four RBPs and four RASEs were validated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and showed high consistency with RNA sequencing (RNA-seq).
Conclusion: Dysregulated RBPs and their associated RASEs may play regulatory roles in MVP development and may therefore be used as therapeutic targets in the future.
Keywords: RNA sequencing; RNA-binding protein; alternative splicing; genome-wide analysis; mitral valve prolapse.
Copyright © 2023 Zhao, Zhou, Tang, Liu, Dai, Xie, Wang, Chen and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Genome-wide identification of dysregulated alternative splicing and RNA-binding proteins involved in atopic dermatitis.Front Genet. 2024 Mar 1;15:1287111. doi: 10.3389/fgene.2024.1287111. eCollection 2024. Front Genet. 2024. PMID: 38495671 Free PMC article.
-
Genome-wide analysis revealed the dysregulation of RNA binding protein-correlated alternative splicing events in myocardial ischemia reperfusion injury.BMC Med Genomics. 2023 Oct 19;16(1):251. doi: 10.1186/s12920-023-01706-5. BMC Med Genomics. 2023. PMID: 37858115 Free PMC article.
-
Identification of alternative splicing and RNA-binding proteins involved in myocardial ischemia-reperfusion injury.Genome. 2023 Oct 1;66(10):261-268. doi: 10.1139/gen-2022-0102. Epub 2023 Jul 19. Genome. 2023. PMID: 37466303
-
Floppy mitral valve (FMV)/mitral valve prolapse (MVP) and the FMV/MVP syndrome: pathophysiologic mechanisms and pathogenesis of symptoms.Cardiology. 2013;126(2):69-80. doi: 10.1159/000351094. Epub 2013 Aug 8. Cardiology. 2013. PMID: 23942374 Review.
-
Genomic analysis in patients with myxomatous mitral valve prolapse: current state of knowledge.BMC Cardiovasc Disord. 2018 Feb 27;18(1):41. doi: 10.1186/s12872-018-0755-y. BMC Cardiovasc Disord. 2018. PMID: 29486707 Free PMC article.
Cited by
-
RNA-binding proteins regulate immune-related alternative splicing in inherited salt-losing tubulopathies.Orphanet J Rare Dis. 2025 Aug 9;20(1):416. doi: 10.1186/s13023-025-03972-1. Orphanet J Rare Dis. 2025. PMID: 40783707 Free PMC article.
References
-
- Constant Dit Beaufils AL, Huttin O, Jobbe-Duval A, Senage T, Filippetti L, Piriou N, et al. . Replacement myocardial fibrosis in patients with mitral valve prolapse: relation to mitral regurgitation, ventricular remodeling, and arrhythmia. Circulation. (2021) 143(18):1763–74. doi: 10.1161/CIRCULATIONAHA.120.050214 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

