Barcode demultiplexing of nanopore sequencing raw signals by unsupervised machine learning
- PMID: 37181486
- PMCID: PMC10173771
- DOI: 10.3389/fbinf.2023.1067113
Barcode demultiplexing of nanopore sequencing raw signals by unsupervised machine learning
Abstract
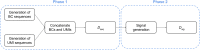
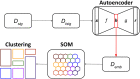
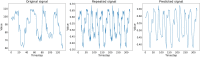
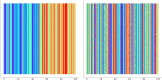
Introduction: Oxford Nanopore Technologies (ONT) is a third generation sequencing approach that allows the analysis of individual, full-length nucleic acids. ONT records the alterations of an ionic current flowing across a nano-scaled pore while a DNA or RNA strand is threading through the pore. Basecalling methods are then leveraged to translate the recorded signal back to the nucleic acid sequence. However, basecall generally introduces errors that hinder the process of barcode demultiplexing, a pivotal task in single-cell RNA sequencing that allows for separating the sequenced transcripts on the basis of their cell of origin. Methods: To solve this issue, we present a novel framework, called UNPLEX, designed to tackle the barcode demultiplexing problem by operating directly on the recorded signals. UNPLEX combines two unsupervised machine learning methods: autoencoders and self-organizing maps (SOM). The autoencoders extract compact, latent representations of the recorded signals that are then clustered by the SOM. Results and Discussion: Our results, obtained on two datasets composed of in silico generated ONT-like signals, show that UNPLEX represents a promising starting point for the development of effective tools to cluster the signals corresponding to the same cell.
Keywords: RNA barcoding; artificial intelligence; autoencoder; complexity reduction; nanopore; scRNA-seq; self-organising map; unsupervised learning.
Copyright © 2023 Papetti, Spolaor, Nazari, Tirelli, Leonardi, Caprioli, Besozzi, Vlachou, Pelicci, Cazzaniga and Nobile.
Conflict of interest statement
TL received reimbursement of expenses from ONT to speak at an event. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Alibrahim H., Ludwig S. A. (2021). “Hyperparameter optimization: Comparing genetic algorithm against grid search and bayesian optimization,” in 2021 IEEE Congress on Evolutionary Computation (CEC) (IEEE; ), 1551–1559.
-
- Breiman L. (2001). Random forests. Mach. Learn. 45, 5–32. 10.1023/a:1010933404324 - DOI
-
- Fowlkes E. B., Mallows C. L. (1983). A method for comparing two hierarchical clusterings. J. Am. Stat. Assoc. 78, 553–569. 10.1080/01621459.1983.10478008 - DOI
LinkOut - more resources
Full Text Sources

