Methylated guanosine and uridine modifications in S. cerevisiae mRNAs modulate translation elongation
- PMID: 37181630
- PMCID: PMC10170649
- DOI: 10.1039/d2cb00229a
Methylated guanosine and uridine modifications in S. cerevisiae mRNAs modulate translation elongation
Abstract
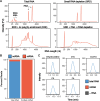
Chemical modifications to protein encoding messenger RNAs (mRNAs) influence their localization, translation, and stability within cells. Over 15 different types of mRNA modifications have been observed by sequencing and liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) approaches. While LC-MS/MS is arguably the most essential tool available for studying analogous protein post-translational modifications, the high-throughput discovery and quantitative characterization of mRNA modifications by LC-MS/MS has been hampered by the difficulty of obtaining sufficient quantities of pure mRNA and limited sensitivities for modified nucleosides. We have overcome these challenges by improving the mRNA purification and LC-MS/MS pipelines. The methodologies we developed result in no detectable non-coding RNA modifications signals in our purified mRNA samples, quantify 50 ribonucleosides in a single analysis, and provide the lowest limit of detection reported for ribonucleoside modification LC-MS/MS analyses. These advancements enabled the detection and quantification of 13 S. cerevisiae mRNA ribonucleoside modifications and reveal the presence of four new S. cerevisiae mRNA modifications at low to moderate levels (1-methyguanosine, N2-methylguanosine, N2,N2-dimethylguanosine, and 5-methyluridine). We identified four enzymes that incorporate these modifications into S. cerevisiae mRNAs (Trm10, Trm11, Trm1, and Trm2, respectively), though our results suggest that guanosine and uridine nucleobases are also non-enzymatically methylated at low levels. Regardless of whether they are incorporated in a programmed manner or as the result of RNA damage, we reasoned that the ribosome will encounter the modifications that we detect in cells. To evaluate this possibility, we used a reconstituted translation system to investigate the consequences of modifications on translation elongation. Our findings demonstrate that the introduction of 1-methyguanosine, N2-methylguanosine and 5-methyluridine into mRNA codons impedes amino acid addition in a position dependent manner. This work expands the repertoire of nucleoside modifications that the ribosome must decode in S. cerevisiae. Additionally, it highlights the challenge of predicting the effect of discrete modified mRNA sites on translation de novo because individual modifications influence translation differently depending on mRNA sequence context.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
-
- Boccaletto P. Machnicka M. A. Purta E. Piątkowski P. Bagiński B. Wirecki T. K. de Crécy-Lagard V. Ross R. Limbach P. A. Kotter A. Helm M. Bujnicki J. M. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–D307. doi: 10.1093/nar/gkx1030. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

