NeuroWRAP: integrating, validating, and sharing neurodata analysis workflows
- PMID: 37181735
- PMCID: PMC10166805
- DOI: 10.3389/fninf.2023.1082111
NeuroWRAP: integrating, validating, and sharing neurodata analysis workflows
Abstract
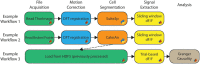
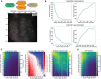
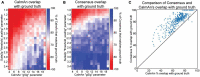
Multiphoton calcium imaging is one of the most powerful tools in modern neuroscience. However, multiphoton data require significant pre-processing of images and post-processing of extracted signals. As a result, many algorithms and pipelines have been developed for the analysis of multiphoton data, particularly two-photon imaging data. Most current studies use one of several algorithms and pipelines that are published and publicly available, and add customized upstream and downstream analysis elements to fit the needs of individual researchers. The vast differences in algorithm choices, parameter settings, pipeline composition, and data sources combine to make collaboration difficult, and raise questions about the reproducibility and robustness of experimental results. We present our solution, called NeuroWRAP (www.neurowrap.org), which is a tool that wraps multiple published algorithms together, and enables integration of custom algorithms. It enables development of collaborative, shareable custom workflows and reproducible data analysis for multiphoton calcium imaging data enabling easy collaboration between researchers. NeuroWRAP implements an approach to evaluate the sensitivity and robustness of the configured pipelines. When this sensitivity analysis is applied to a crucial step of image analysis, cell segmentation, we find a substantial difference between two popular workflows, CaImAn and Suite2p. NeuroWRAP harnesses this difference by introducing consensus analysis, utilizing two workflows in conjunction to significantly increase the trustworthiness and robustness of cell segmentation results.
Keywords: consensus; image analysis; reproducibility; two-photon calcium imaging; workflow management.
Copyright © 2023 Bowen, Magnusson, Diep, Ayyangar, Smirnov, Kanold and Losert.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials

