Isoamphipathic antibacterial molecules regulating activity and toxicity through positional isomerism
- PMID: 37181778
- PMCID: PMC10171078
- DOI: 10.1039/d2sc06065e
Isoamphipathic antibacterial molecules regulating activity and toxicity through positional isomerism
Abstract
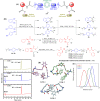
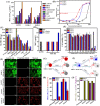
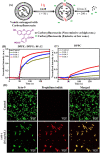
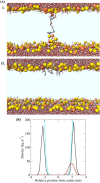
Peptidomimetic antimicrobials exhibit a selective interaction with bacterial cells over mammalian cells once they have achieved an optimum amphiphilic balance (hydrophobicity/hydrophilicity) in the molecular architecture. To date, hydrophobicity and cationic charge have been considered the crucial parameters to attain such amphiphilic balance. However, optimization of these properties is not enough to circumvent unwanted toxicity towards mammalian cells. Hence, herein, we report new isoamphipathic antibacterial molecules (IAMs: 1-3) where positional isomerism was introduced as one of the guiding factors for molecular design. This class of molecules displayed good (MIC = 1-8 μg mL-1 or μM) to moderate [MIC = 32-64 μg mL-1 (32.2-64.4 μM)] antibacterial activity against multiple Gram-positive and Gram-negative bacteria. Positional isomerism showed a strong influence on regulating antibacterial activity and toxicity for ortho [IAM-1: MIC = 1-32 μg mL-1 (1-32.2 μM), HC50 = 650 μg mL-1 (654.6 μM)], meta [IAM-2: MIC = 1-16 μg mL-1 (1-16.1 μM), HC50 = 98 μg mL-1 (98.7 μM)] and para [IAM-3: MIC = 1-16 μg mL-1 (1-16.1 μM), HC50 = 160 μg mL-1 (161.1 μM)] isomers. Co-culture studies and investigation of membrane dynamics indicated that ortho isomer, IAM-1 exerted more selective activity towards bacterial over mammalian membranes, compared to meta and para isomers. Furthermore, the mechanism of action of the lead molecule (IAM-1) has been characterized through detailed molecular dynamics simulations. In addition, the lead molecule displayed substantial efficacy against dormant bacteria and mature biofilms, unlike conventional antibiotics. Importantly, IAM-1 exhibited moderate in vivo activity against MRSA wound infection in a murine model with no detectable dermal toxicity. Altogether, the report explored the design and development of isoamphipathic antibacterial molecules to establish the role of positional isomerism in achieving selective and potential antibacterial agents.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
LinkOut - more resources
Full Text Sources

