Full-spectrum HIV drug resistance mutation detection by high-resolution complete pol gene sequencing
- PMID: 37182384
- PMCID: PMC10330399
- DOI: 10.1016/j.jcv.2023.105491
Full-spectrum HIV drug resistance mutation detection by high-resolution complete pol gene sequencing
Abstract
Background: Drug resistance mutation testing is a key element for HIV clinical management, informing effective treatment regimens. However, resistance screening in current clinical practice is limited in reporting linked cross-class resistance mutations and minority variants, both of which may increase the risk of virological failure.
Methods: To address these limitations, we obtained 358 full-length pol gene sequences from 52 specimens of 20 HIV infected individuals by combining microdroplet amplification, unique molecular identifier (UMI) labeling, and long-read high-throughput sequencing.
Results: We conducted a rigorous assessment of the accuracy of our pipeline for precision drug resistance mutation detection, verifying that a sequencing depth of 35 high-throughput reads achieved complete, error-free pol gene sequencing. We detected 26 distinct drug resistance mutations to Protease Inhibitors (PIs), Nucleoside Reverse Transcriptase Inhibitors (NRTIs), Non-Nucleoside Reverse Transcriptase Inhibitors (NNRTIs), and Integrase Strand Transfer Inhibitors (INSTIs). We detected linked cross-class drug resistance mutations (PI+NRTI, PI+NNRTI, and NRTI+NNRTI) that confer cross-resistance to multiple drugs in different classes. Fourteen different types of minority mutations were also detected with frequencies ranging from 3.2% to 19%, and the presence of these mutations was verified by Sanger reference sequencing. We detected a putative transmitted drug resistance mutation (TDRM) in one individual that persisted for over seven months from the first sample collected at the acute stage of infection prior to seroconversion.
Conclusions: Our comprehensive drug resistance mutation profiling can advance clinical practice by reporting mutation linkage and minority variants to better guide antiretroviral therapy options.
Keywords: Cross-resistance; Drug resistance mutations; HIV; Minority variants.
Copyright © 2023 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

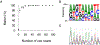



References
-
- UNAIDS. Global HIV & AIDS statistics - Fact sheet. 2021.
-
- Cozzi-Lepri A, Noguera-Julian M, Di Giallonardo F, Schuurman R, Daumer M, Aitken S, et al. Low-frequency drug-resistant HIV-1 and risk of virological failure to first-line NNRTI-based ART: a multicohort European case-control study using centralized ultrasensitive 454 pyrosequencing. J Antimicrob Chemother. 2015;70(3):930–40. - PMC - PubMed
-
- Geretti AM, Fox ZV, Booth CL, Smith CJ, Phillips AN, Johnson M, et al. Low-frequency K103N strengthens the impact of transmitted drug resistance on virologic responses to first-line efavirenz or nevirapine-based highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;52(5):569–73. - PubMed
-
- Goodman DD, Zhou Y, Margot NA, McColl DJ, Zhong L, Borroto-Esoda K, et al. Low level of the K103N HIV-1 above a threshold is associated with virological failure in treatment-naive individuals undergoing efavirenz-containing therapy. AIDS. 2011;25(3):325–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

