Genotype-phenotype correlation in contactin-associated protein-like 2 (CNTNAP-2) developmental disorder
- PMID: 37183190
- PMCID: PMC10329570
- DOI: 10.1007/s00439-023-02552-2
Genotype-phenotype correlation in contactin-associated protein-like 2 (CNTNAP-2) developmental disorder
Abstract
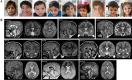
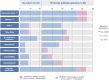
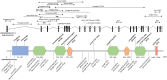
Contactin-associated protein-like 2 (CNTNAP2) gene encodes for CASPR2, a presynaptic type 1 transmembrane protein, involved in cell-cell adhesion and synaptic interactions. Biallelic CNTNAP2 loss has been associated with "Pitt-Hopkins-like syndrome-1" (MIM#610042), while the pathogenic role of heterozygous variants remains controversial. We report 22 novel patients harboring mono- (n = 2) and bi-allelic (n = 20) CNTNAP2 variants and carried out a literature review to characterize the genotype-phenotype correlation. Patients (M:F 14:8) were aged between 3 and 19 years and affected by global developmental delay (GDD) (n = 21), moderate to profound intellectual disability (n = 17) and epilepsy (n = 21). Seizures mainly started in the first two years of life (median 22.5 months). Antiseizure medications were successful in controlling the seizures in about two-thirds of the patients. Autism spectrum disorder (ASD) and/or other neuropsychiatric comorbidities were present in nine patients (40.9%). Nonspecific midline brain anomalies were noted in most patients while focal signal abnormalities in the temporal lobes were noted in three subjects. Genotype-phenotype correlation was performed by also including 50 previously published patients (15 mono- and 35 bi-allelic variants). Overall, GDD (p < 0.0001), epilepsy (p < 0.0001), hyporeflexia (p = 0.012), ASD (p = 0.009), language impairment (p = 0.020) and severe cognitive impairment (p = 0.031) were significantly associated with the presence of biallelic versus monoallelic variants. We have defined the main features associated with biallelic CNTNAP2 variants, as severe cognitive impairment, epilepsy and behavioral abnormalities. We propose CASPR2-deficiency neurodevelopmental disorder as an exclusively recessive disease while the contribution of heterozygous variants is less likely to follow an autosomal dominant inheritance pattern.
© 2023. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures

References
-
- Alarcón M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, Nelson SF, Cantor RM, Geschwind DH. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82(1):150–159. doi: 10.1016/j.ajhg.2007.09.005. - DOI - PMC - PubMed
-
- Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Chakravarti A. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82(1):160–164. doi: 10.1016/j.ajhg.2007.09.015. - DOI - PMC - PubMed
-
- Badshah N, Mattison KA, Ahmad S, Chopra P, Johnston HR, Ahmad S, Khan SH, Sarwar MT, Cutler DJ, Taylor M, Vadlamani G, Zwick ME, Escayg A. Novel missense CNTNAP2 variant identified in two consanguineous Pakistani families with developmental delay, epilepsy, intellectual disability, and aggressive behavior. Front Neurol. 2022;13:918022. doi: 10.3389/fneur.2022.918022. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

