Tender love and disassembly: How a TLDc domain protein breaks the V-ATPase
- PMID: 37183929
- PMCID: PMC10392918
- DOI: 10.1002/bies.202200251
Tender love and disassembly: How a TLDc domain protein breaks the V-ATPase
Abstract
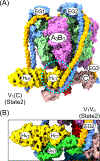
Vacuolar ATPases (V-ATPases, V1 Vo -ATPases) are rotary motor proton pumps that acidify intracellular compartments, and, when localized to the plasma membrane, the extracellular space. V-ATPase is regulated by a unique process referred to as reversible disassembly, wherein V1 -ATPase disengages from Vo proton channel in response to diverse environmental signals. Whereas the disassembly step of this process is ATP dependent, the (re)assembly step is not, but requires the action of a heterotrimeric chaperone referred to as the RAVE complex. Recently, an alternative pathway of holoenzyme disassembly was discovered that involves binding of Oxidation Resistance 1 (Oxr1p), a poorly characterized protein implicated in oxidative stress response. Unlike conventional reversible disassembly, which depends on enzyme activity, Oxr1p induced dissociation can occur in absence of ATP. Yeast Oxr1p belongs to the family of TLDc domain containing proteins that are conserved from yeast to mammals, and have been implicated in V-ATPase function in a variety of tissues. This brief perspective summarizes what we know about the molecular mechanisms governing both reversible (ATP dependent) and Oxr1p driven (ATP independent) V-ATPase dissociation into autoinhibited V1 and Vo subcomplexes.
Keywords: ADP inhibition; Oxr1p; TLDc domain; V1-ATPase; Vo proton channel; peripheral stalk; protein-protein interaction; reversible disassembly; subunit C; subunit H; vacuolar H+-ATPase.
© 2023 The Authors. BioEssays published by Wiley Periodicals LLC.
Conflict of interest statement
Conflict of interest
The authors declare that they have no conflict of interest.
Figures




References
-
- Maxson ME, Grinstein S. 2014. The vacuolar-type H(+)-ATPase at a glance - more than a proton pump. J Cell Sci 127: 4987–93. - PubMed
-
- Santos-Pereira C, Rodrigues LR, Côrte-Real M. 2021. Emerging insights on the role of V-ATPase in human diseases: Therapeutic challenges and opportunities. Med Res Rev. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

