Domain tethering impacts dimerization and DNA-mediated allostery in the human transcription factor FoxP1
- PMID: 37184020
- PMCID: PMC10188207
- DOI: 10.1063/5.0138782
Domain tethering impacts dimerization and DNA-mediated allostery in the human transcription factor FoxP1
Abstract
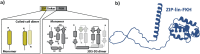
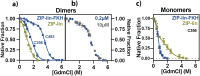
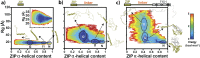
Transcription factors are multidomain proteins with specific DNA binding and regulatory domains. In the human FoxP subfamily (FoxP1, FoxP2, FoxP3, and FoxP4) of transcription factors, a 90 residue-long disordered region links a Leucine Zipper (ZIP)-known to form coiled-coil dimers-and a Forkhead (FKH) domain-known to form domain swapping dimers. We used replica exchange discrete molecular dynamics simulations, single-molecule fluorescence experiments, and other biophysical tools to understand how domain tethering in FoxP1 impacts dimerization at ZIP and FKH domains and how DNA binding allosterically regulates their dimerization. We found that domain tethering promotes FoxP1 dimerization but inhibits a FKH domain-swapped structure. Furthermore, our findings indicate that the linker mediates the mutual organization and dynamics of ZIP and FKH domains, forming closed and open states with and without interdomain contacts, thus highlighting the role of the linkers in multidomain proteins. Finally, we found that DNA allosterically promotes structural changes that decrease the dimerization propensity of FoxP1. We postulate that, upon DNA binding, the interdomain linker plays a crucial role in the gene regulatory function of FoxP1.
© 2023 Author(s). All article content, except where otherwise noted, is licensed under a Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Conflict of interest statement
The authors have no conflicts to disclose.
Figures


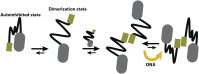
References
-
- Damante G., Fabbro D., Pellizzari L., Civitareale D., Guazzi S., Polycarpou-Schwartz M., Cauci S., Quadrifoglio F., Formisano S., and Lauro R. D., “Sequence-specific DNA recognition by the thyroid transcription factor-1 homeodomain,” Nucleic Acids Res. 22(15), 3075–3083 (1994).10.1093/nar/22.15.3075 - DOI - PMC - PubMed
-
- Rastogi C., Rube H. T., Kribelbauer J. F., Crocker J., Loker R. E., Martini G. D., Laptenko O., Freed-Pastor W. A., Prives C., Stern D. L., Mann R. S., and Bussemaker H. J., “Accurate and sensitive quantification of protein-DNA binding affinity,” Proc. Natl. Acad. Sci. U. S. A. 115(16), E3692 (2018).10.1073/pnas.1714376115 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

