PhAT: A Flexible Open-Source GUI-Driven Toolkit for Photometry Analysis
- PMID: 37184156
- PMCID: PMC10246504
- DOI: 10.1002/cpz1.763
PhAT: A Flexible Open-Source GUI-Driven Toolkit for Photometry Analysis
Abstract
Photometry approaches detect sensor-mediated changes in fluorescence as a proxy for rapid molecular changes within the brain. As a flexible technique with a relatively low cost to implement, photometry is rapidly being incorporated into neuroscience laboratories. Yet, although multiple data acquisition systems for photometry now exist, robust analytical pipelines for the resulting data remain limited. Here we present the Photometry Analysis Toolkit (PhAT)-a free open-source analysis pipeline that provides options for signal normalization, incorporation of multiple data streams to align photometry data with behavior and other events, calculation of event-related changes in fluorescence, and comparison of similarity across fluorescent traces. A graphical user interface (GUI) enables use of this software without prior coding knowledge. In addition to providing foundational analytical tools, PhAT is designed to readily incorporate community-driven development of new modules for more bespoke analyses, and enables data to be easily exported to enable subsequent statistical testing and/or code-based analyses. In addition, we provide recommendations regarding technical aspects of photometry experiments, including sensor selection and validation, reference signal considerations, and best practices for experimental design and data collection. We hope that the distribution of this software and protocols will lower the barrier to entry for new photometry users and improve the quality of collected data, increasing transparency and reproducibility in photometry analyses. © 2023 Wiley Periodicals LLC. Basic Protocol 1: Software and environment installation Alternate Protocol 1: Software and environment update Basic Protocol 2: GUI-driven fiber photometry analysis Support Protocol 1: Examining signal quality Support Protocol 2: Interacting with graphs Basic Protocol 3: Adding modules to PhAT Alternate Protocol 2: Creating functions for use in Jupyter Notebook.
Keywords: analysis; open-source; photometry; software.
© 2023 Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT:
Authors declare no conflicts of interest.
Figures




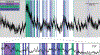
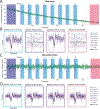
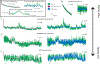


Update of
-
PhAT: A flexible open-source GUI-driven toolkit for photometry analysis.bioRxiv [Preprint]. 2023 Mar 14:2023.03.14.532489. doi: 10.1101/2023.03.14.532489. bioRxiv. 2023. Update in: Curr Protoc. 2023 May;3(5):e763. doi: 10.1002/cpz1.763. PMID: 36993180 Free PMC article. Updated. Preprint.
References
LITERATURE CITED:
-
- Adelsberger H, Garaschuk O, and Konnerth A 2005. Cortical calcium waves in resting newborn mice. Nature Neuroscience 8:988–990. - PubMed
INTERNET RESOURCES:
-
- To access our code base visit: https://github.com/donaldsonlab/PhAT
-
- For information on how to install python and relevant download links visit: https://www.python.org/downloads/ or https://wiki.python.org/moin/BeginnersGUIde/Download
-
- For information on how to install anaconda and the relevant download links visit: https://docs.anaconda.com/anaconda/install/
-
- For information on how to install pip and the relevant download links visit: https://pip.pypa.io/en/stable/installation/
-
- For information and tutorials on how to use jupyterlab or jupyter notebook visit: https://www.tutorialspoint.com/jupyter/jupyterlab_overview.htm or https://www.tutorialspoint.com/jupyter/jupyter_notebook_introduction.htm
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

