Designed Rubredoxin miniature in a fully artificial electron chain triggered by visible light
- PMID: 37185349
- PMCID: PMC10130062
- DOI: 10.1038/s41467-023-37941-8
Designed Rubredoxin miniature in a fully artificial electron chain triggered by visible light
Abstract
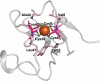
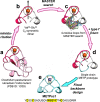
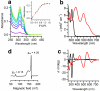
Designing metal sites into de novo proteins has significantly improved, recently. However, identifying the minimal coordination spheres, able to encompass the necessary information for metal binding and activity, still represents a great challenge, today. Here, we test our understanding with a benchmark, nevertheless difficult, case. We assemble into a miniature 28-residue protein, the quintessential elements required to fold properly around a FeCys4 redox center, and to function efficiently in electron-transfer. This study addresses a challenge in de novo protein design, as it reports the crystal structure of a designed tetra-thiolate metal-binding protein in sub-Å agreement with the intended design. This allows us to well correlate structure to spectroscopic and electrochemical properties. Given its high reduction potential compared to natural and designed FeCys4-containing proteins, we exploit it as terminal electron acceptor of a fully artificial chain triggered by visible light.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Tang, K.-H. & Blankenship, R. E. Photosynthetic Electron Transport. in Encyclopedia of Biophysics (ed. Roberts, G. C. K.) 1868–1873 (Springer Berlin Heidelberg). 10.1007/978-3-642-16712-6_20 (2013).
Grants and funding
- SEA-WAVE 2020BKK3W9, [CUP_E69J22001140005]/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- SEA-WAVE 2020BKK3W9, [CUP_E69J22001140005]/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- Programma Operativo FESR Campania 2014-2020, Asse 1, [CUP B63D18000350007]/Regione Campania
- 813209 (PARACAT)/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Skłodowska-Curie Actions (H2020 Excellent Science - Marie Skłodowska-Curie Actions)
- 813209 (PARACAT)/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Skłodowska-Curie Actions (H2020 Excellent Science - Marie Skłodowska-Curie Actions)
LinkOut - more resources
Full Text Sources

