Applications and Tuning Strategies for Transcription Factor-Based Metabolite Biosensors
- PMID: 37185503
- PMCID: PMC10136082
- DOI: 10.3390/bios13040428
Applications and Tuning Strategies for Transcription Factor-Based Metabolite Biosensors
Abstract
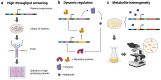
Transcription factor (TF)-based biosensors are widely used for the detection of metabolites and the regulation of cellular pathways in response to metabolites. Several challenges hinder the direct application of TF-based sensors to new hosts or metabolic pathways, which often requires extensive tuning to achieve the optimal performance. These tuning strategies can involve transcriptional or translational control depending on the parameter of interest. In this review, we highlight recent strategies for engineering TF-based biosensors to obtain the desired performance and discuss additional design considerations that may influence a biosensor's performance. We also examine applications of these sensors and suggest important areas for further work to continue the advancement of small-molecule biosensors.
Keywords: biosensor applications; biosensor tuning; dynamic regulation; high-throughput screening; metabolic heterogeneity; transcription factor; transcriptional control; translational control.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


Similar articles
-
[Progress in transcription factor-based metabolite biosensors].Sheng Wu Gong Cheng Xue Bao. 2021 Mar 25;37(3):911-922. doi: 10.13345/j.cjb.200641. Sheng Wu Gong Cheng Xue Bao. 2021. PMID: 33783157 Review. Chinese.
-
Advances in engineering and optimization of transcription factor-based biosensors for plug-and-play small molecule detection.Curr Opin Biotechnol. 2022 Aug;76:102753. doi: 10.1016/j.copbio.2022.102753. Epub 2022 Jul 21. Curr Opin Biotechnol. 2022. PMID: 35872379 Review.
-
The Growth Dependent Design Constraints of Transcription-Factor-Based Metabolite Biosensors.ACS Synth Biol. 2022 Jul 15;11(7):2247-2258. doi: 10.1021/acssynbio.2c00143. Epub 2022 Jun 14. ACS Synth Biol. 2022. PMID: 35700119 Free PMC article.
-
Applications and advances of metabolite biosensors for metabolic engineering.Metab Eng. 2015 Sep;31:35-43. doi: 10.1016/j.ymben.2015.06.008. Epub 2015 Jul 2. Metab Eng. 2015. PMID: 26142692 Review.
-
Genetically Encoded Biosensor Engineering for Application in Directed Evolution.J Microbiol Biotechnol. 2023 Oct 28;33(10):1257-1267. doi: 10.4014/jmb.2304.04031. Epub 2023 Jul 14. J Microbiol Biotechnol. 2023. PMID: 37449325 Free PMC article. Review.
Cited by
-
Biological Switches: Past and Future Milestones of Transcription Factor-Based Biosensors.ACS Synth Biol. 2025 Jan 17;14(1):72-86. doi: 10.1021/acssynbio.4c00689. Epub 2024 Dec 22. ACS Synth Biol. 2025. PMID: 39709556 Review.
-
Diverse mechanisms of bioproduction heterogeneity in fermentation and their control strategies.J Ind Microbiol Biotechnol. 2023 Feb 17;50(1):kuad033. doi: 10.1093/jimb/kuad033. J Ind Microbiol Biotechnol. 2023. PMID: 37791393 Free PMC article. Review.
-
Establishing a Malonyl-CoA Biosensor for the Two Model Cyanobacteria Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942.ACS Synth Biol. 2025 Jul 18;14(7):2865-2877. doi: 10.1021/acssynbio.5c00320. Epub 2025 Jun 30. ACS Synth Biol. 2025. PMID: 40588753 Free PMC article.
-
High throughput construction of species characterized bacterial biobank for functional bacteria screening: demonstration with GABA-producing bacteria.Front Microbiol. 2025 Mar 27;16:1545877. doi: 10.3389/fmicb.2025.1545877. eCollection 2025. Front Microbiol. 2025. PMID: 40212379 Free PMC article.
-
Enhancing glucaric acid production from myo-inositol in Escherichia coli by eliminating cell-to-cell variation.Appl Environ Microbiol. 2024 Jun 18;90(6):e0014924. doi: 10.1128/aem.00149-24. Epub 2024 May 29. Appl Environ Microbiol. 2024. PMID: 38808978 Free PMC article.
References
-
- Schroeder G.M., Akinyemi O., Malik J., Focht C.M., Pritchett E.M., Baker C.D., McSally J.P., Jenkins J.L., Mathews D.H., Wedekind J.E. A Riboswitch Separated from Its Ribosome-Binding Site Still Regulates Translation. Nucleic Acids Res. 2023;51:2464–2484. doi: 10.1093/nar/gkad056. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

