Bioinformatic survey of CRISPR loci across 15 Serratia species
- PMID: 37186230
- PMCID: PMC9981886
- DOI: 10.1002/mbo3.1339
Bioinformatic survey of CRISPR loci across 15 Serratia species
Abstract
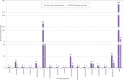
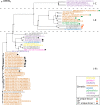
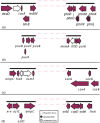
The Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR-associated proteins (CRISPR-Cas) system of prokaryotes is an adaptative immune defense mechanism to protect themselves from invading genetic elements (e.g., phages and plasmids). Studies that describe the genetic organization of these prokaryotic systems have mainly reported on the Enterobacteriaceae family (now reorganized within the order of Enterobacterales). For some genera, data on CRISPR-Cas systems remain poor, as in the case of Serratia (now part of the Yersiniaceae family) where data are limited to a few genomes of the species marcescens. This study describes the detection, in silico, of CRISPR loci in 146 Serratia complete genomes and 336 high-quality assemblies available for the species ficaria, fonticola, grimesii, inhibens, liquefaciens, marcescens, nematodiphila, odorifera, oryzae, plymuthica, proteomaculans, quinivorans, rubidaea, symbiotica, and ureilytica. Apart from subtypes I-E and I-F1 which had previously been identified in marcescens, we report that of I-C and the I-E unique locus 1, I-E*, and I-F1 unique locus 1. Analysis of the genomic contexts for CRISPR loci revealed mdtN-phnP as the region mostly shared (grimesii, inhibens, marcescens, nematodiphila, plymuthica, rubidaea, and Serratia sp.). Three new contexts detected in genomes of rubidaea and fonticola (puu genes-mnmA) and rubidaea (osmE-soxG and ampC-yebZ) were also found. The plasmid and/or phage origin of spacers was also established.
Keywords: CRISPR system; RPW; Rhynchophorus ferrugineus; subtype I-C; subtype I-E; subtype I-F1.
© 2022 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures



References
-
- Adeolu, M. , Alnajar, S. , Naushad, S. , & S Gupta, R. (2016). Genome‐based phylogeny and taxonomy of the “enterobacteriales”: Proposal for enterobacterales ord. nov. divided into the families enterobacteriaceae, erwiniaceae fam. nov., pectobacteriaceae fam. nov., yersiniaceae fam. nov., hafniaceae fam. nov., morganellaceae fam. nov., and budviciaceae fam. nov. International Journal of Systematic and Evolutionary Microbiology, 66(12), 5575–5599. 10.1099/ijsem.0.001485 - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

