Receptor for hyaluronan-mediated motility (RHAMM) defines an invasive niche associated with tumor progression and predicts poor outcomes in breast cancer patients
- PMID: 37186300
- PMCID: PMC10417882
- DOI: 10.1002/path.6082
Receptor for hyaluronan-mediated motility (RHAMM) defines an invasive niche associated with tumor progression and predicts poor outcomes in breast cancer patients
Abstract
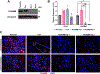
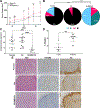
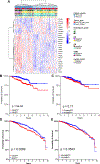
Breast cancer invasion and metastasis result from a complex interplay between tumor cells and the tumor microenvironment (TME). Key oncogenic changes in the TME include aberrant synthesis, processing, and signaling of hyaluronan (HA). Hyaluronan-mediated motility receptor (RHAMM, CD168; HMMR) is an HA receptor enabling tumor cells to sense and respond to this aberrant TME during breast cancer progression. Previous studies have associated RHAMM expression with breast tumor progression; however, cause and effect mechanisms are incompletely established. Focused gene expression analysis of an internal breast cancer patient cohort confirmed that increased RHAMM expression correlates with aggressive clinicopathological features. To probe mechanisms, we developed a novel 27-gene RHAMM-related signature (RRS) by intersecting differentially expressed genes in lymph node (LN)-positive patient cases with the transcriptome of a RHAMM-dependent model of cell transformation, which we validated in an independent cohort. We demonstrate that the RRS predicts for poor survival and is enriched for cell cycle and TME-interaction pathways. Further analyses using CRISPR/Cas9-generated RHAMM-/- breast cancer cells provided direct evidence that RHAMM promotes invasion in vitro and in vivo. Immunohistochemistry studies highlighted heterogeneous RHAMM protein expression, and spatial transcriptomics associated the RRS with RHAMM-high microanatomic foci. We conclude that RHAMM upregulation leads to the formation of 'invasive niches', which are enriched in RRS-related pathways that drive invasion and could be targeted to limit invasive progression and improve patient outcomes. © 2023 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
Keywords: breast cancer; cell division; extracellular matrix; hyaluronan; hyaluronan receptors; neoplasm invasiveness; tumor microenvironment; xenograft.
© 2023 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.
Conflict of interest statement
No conflicts of interest were declared.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

