Development of early life gut resistome and mobilome across gestational ages and microbiota-modifying treatments
- PMID: 37187112
- PMCID: PMC10192547
- DOI: 10.1016/j.ebiom.2023.104613
Development of early life gut resistome and mobilome across gestational ages and microbiota-modifying treatments
Abstract
Background: Gestational age (GA) and associated level of gastrointestinal tract maturation are major factors driving the initial gut microbiota composition in preterm infants. Besides, compared to term infants, premature infants often receive antibiotics to treat infections and probiotics to restore optimal gut microbiota. How GA, antibiotics, and probiotics modulate the microbiota's core characteristics, gut resistome and mobilome, remains nascent.
Methods: We analysed metagenomic data from a longitudinal observational study in six Norwegian neonatal intensive care units to describe the bacterial microbiota of infants of varying GA and receiving different treatments. The cohort consisted of probiotic-supplemented and antibiotic-exposed extremely preterm infants (n = 29), antibiotic-exposed very preterm (n = 25), antibiotic-unexposed very preterm (n = 8), and antibiotic-unexposed full-term (n = 10) infants. The stool samples were collected on days of life 7, 28, 120, and 365, and DNA extraction was followed by shotgun metagenome sequencing and bioinformatical analysis.
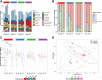
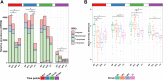
Findings: The top predictors of microbiota maturation were hospitalisation length and GA. Probiotic administration rendered the gut microbiota and resistome of extremely preterm infants more alike to term infants on day 7 and ameliorated GA-driven loss of microbiota interconnectivity and stability. GA, hospitalisation, and both microbiota-modifying treatments (antibiotics and probiotics) contributed to an elevated carriage of mobile genetic elements in preterm infants compared to term controls. Finally, Escherichia coli was associated with the highest number of antibiotic-resistance genes, followed by Klebsiella pneumoniae and Klebsiella aerogenes.
Interpretation: Prolonged hospitalisation, antibiotics, and probiotic intervention contribute to dynamic alterations in resistome and mobilome, gut microbiota characteristics relevant to infection risk.
Funding: Odd-Berg Group, Northern Norway Regional Health Authority.
Keywords: Extremely preterm infants; Gestational age; Gut microbiota; Mobilome; Probiotics; Resistome.
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors have declared that no competing interest exists.
Figures





References
-
- Walani S.R. Global burden of preterm birth. Int J Gynaecol Obstet. 2020;150(1):31–33. - PubMed
-
- Kullberg R.F., Wiersinga W.J., Haak B.W. Gut microbiota and sepsis: from pathogenesis to novel treatments. Curr Opin Gastroenterol. 2021;37(6):578–585. - PubMed
-
- Healy D.B., Ryan C.A., Ross R.P., Stanton C., Dempsey E.M. Clinical implications of preterm infant gut microbiome development. Nat Microbiol. 2022;7(1):22–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

