Tumor heterogeneity: preclinical models, emerging technologies, and future applications
- PMID: 37188201
- PMCID: PMC10175698
- DOI: 10.3389/fonc.2023.1164535
Tumor heterogeneity: preclinical models, emerging technologies, and future applications
Abstract
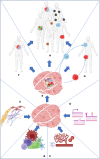
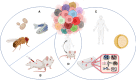
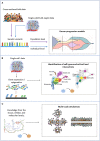
Heterogeneity describes the differences among cancer cells within and between tumors. It refers to cancer cells describing variations in morphology, transcriptional profiles, metabolism, and metastatic potential. More recently, the field has included the characterization of the tumor immune microenvironment and the depiction of the dynamics underlying the cellular interactions promoting the tumor ecosystem evolution. Heterogeneity has been found in most tumors representing one of the most challenging behaviors in cancer ecosystems. As one of the critical factors impairing the long-term efficacy of solid tumor therapy, heterogeneity leads to tumor resistance, more aggressive metastasizing, and recurrence. We review the role of the main models and the emerging single-cell and spatial genomic technologies in our understanding of tumor heterogeneity, its contribution to lethal cancer outcomes, and the physiological challenges to consider in designing cancer therapies. We highlight how tumor cells dynamically evolve because of the interactions within the tumor immune microenvironment and how to leverage this to unleash immune recognition through immunotherapy. A multidisciplinary approach grounded in novel bioinformatic and computational tools will allow reaching the integrated, multilayered knowledge of tumor heterogeneity required to implement personalized, more efficient therapies urgently required for cancer patients.
Keywords: heterogeneity models; human in vitro models; tumor heterogeneity; tumor immune microenvironment; tumor microenvironment (TME).
Copyright © 2023 Proietto, Crippa, Damiani, Pasquale, Sacco, Vanoni and Gilardi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Kashyap A, Rapsomaniki MA, Barros V, Fomitcheva-Khartchenko A, Martinelli AL, Rodriguez AF, et al. Quantification of tumor heterogeneity: from data acquisition to metric generation. Trends Biotechnol (2022) 40(6):647–76. http://www.cell.com/article/S0167779921002675/fulltext. doi: 10.1016/j.tibtech.2021.11.006 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

