A single-cell map of antisense oligonucleotide activity in the brain
- PMID: 37188501
- PMCID: PMC10415122
- DOI: 10.1093/nar/gkad371
A single-cell map of antisense oligonucleotide activity in the brain
Abstract
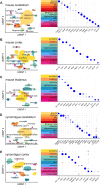
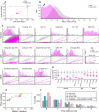
Antisense oligonucleotides (ASOs) dosed into cerebrospinal fluid (CSF) distribute broadly throughout the central nervous system (CNS). By modulating RNA, they hold the promise of targeting root molecular causes of disease and hold potential to treat myriad CNS disorders. Realization of this potential requires that ASOs must be active in the disease-relevant cells, and ideally, that monitorable biomarkers also reflect ASO activity in these cells. The biodistribution and activity of such centrally delivered ASOs have been deeply characterized in rodent and non-human primate (NHP) models, but usually only in bulk tissue, limiting our understanding of the distribution of ASO activity across individual cells and across diverse CNS cell types. Moreover, in human clinical trials, target engagement is usually monitorable only in a single compartment, CSF. We sought a deeper understanding of how individual cells and cell types contribute to bulk tissue signal in the CNS, and how these are linked to CSF biomarker outcomes. We employed single nucleus transcriptomics on tissue from mice treated with RNase H1 ASOs against Prnp and Malat1 and NHPs treated with an ASO against PRNP. Pharmacologic activity was observed in every cell type, though sometimes with substantial differences in magnitude. Single cell RNA count distributions implied target RNA suppression in every single sequenced cell, rather than intense knockdown in only some cells. Duration of action up to 12 weeks post-dose differed across cell types, being shorter in microglia than in neurons. Suppression in neurons was generally similar to, or more robust than, the bulk tissue. In macaques, PrP in CSF was lowered 40% in conjunction with PRNP knockdown across all cell types including neurons, arguing that a CSF biomarker readout is likely to reflect ASO pharmacodynamic effect in disease-relevant cells in a neuronal disorder. Our results provide a reference dataset for ASO activity distribution in the CNS and establish single nucleus sequencing as a method for evaluating cell type specificity of oligonucleotide therapeutics and other modalities.
Plain language summary
Antisense oligonucleotide (ASO) drugs are a type of chemically modified DNA that can be injected into cerebrospinal fluid in order to enter brain cells and reduce the amount of RNA from a specific gene. The brain is a complex mixture of hundreds of billions of cells. When an ASO lowers a target gene's RNA by 50%, is that a 50% reduction in 100% of cells, or a 100% reduction in 50% of cells? Are the many different cell types of the brain affected equally? This new study uses single cell RNA sequencing to answer these questions, finding that ASOs are broadly active across cell types and individual cells, and linking reduction of target protein in cerebrospinal fluid to disease-relevant cells.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Update of
-
A single-cell map of antisense oligonucleotide activity in the brain.bioRxiv [Preprint]. 2023 Feb 14:2023.02.14.528473. doi: 10.1101/2023.02.14.528473. bioRxiv. 2023. Update in: Nucleic Acids Res. 2023 Aug 11;51(14):7109-7124. doi: 10.1093/nar/gkad371. PMID: 36824749 Free PMC article. Updated. Preprint.
References
-
- Crooke S.T., Baker B.F., Crooke R.M., Liang X.-H.. Antisense technology: an overview and prospectus. Nat. Rev. Drug Discov. 2021; 20:427–453. - PubMed
-
- Crooke S.T., Wang S., Vickers T.A., Shen W., Liang X.-H.. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017; 35:230–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

