GalNAc-Lipid nanoparticles enable non-LDLR dependent hepatic delivery of a CRISPR base editing therapy
- PMID: 37188660
- PMCID: PMC10185539
- DOI: 10.1038/s41467-023-37465-1
GalNAc-Lipid nanoparticles enable non-LDLR dependent hepatic delivery of a CRISPR base editing therapy
Abstract
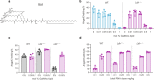
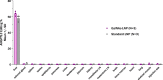
Lipid nanoparticles have demonstrated utility in hepatic delivery of a range of therapeutic modalities and typically deliver their cargo via low-density lipoprotein receptor-mediated endocytosis. For patients lacking sufficient low-density lipoprotein receptor activity, such as those with homozygous familial hypercholesterolemia, an alternate strategy is needed. Here we show the use of structure-guided rational design in a series of mouse and non-human primate studies to optimize a GalNAc-Lipid nanoparticle that allows for low-density lipoprotein receptor independent delivery. In low-density lipoprotein receptor-deficient non-human primates administered a CRISPR base editing therapy targeting the ANGPTL3 gene, the introduction of an optimized GalNAc-based asialoglycoprotein receptor ligand to the nanoparticle surface increased liver editing from 5% to 61% with minimal editing in nontargeted tissues. Similar editing was noted in wild-type monkeys, with durable blood ANGPTL3 protein reduction up to 89% six months post dosing. These results suggest that GalNAc-Lipid nanoparticles may effectively deliver to both patients with intact low-density lipoprotein receptor activity as well as those afflicted by homozygous familial hypercholesterolemia.
© 2023. The Author(s).
Conflict of interest statement
A.V.K. is an employee and holds equity in Verve Therapeutics and has served as a scientific advisor to Amgen, Novartis, Silence Therapeutics, Korro Bio, Veritas International, Color Health, Third Rock Ventures, Illumina, Foresite Labs, and Ambry. K.M. is an advisor to and holds equity in Verve Therapeutics and Variant Bio. S.K. is an employee and officer of Verve Therapeutics, holds equity in Verve Therapeutics, Maze Therapeutics and Relay Therapeutics, and serves on the Board of Directors of Verve Therapeutics, Maze Therapeutics, and Relay Therapeutics. A.M.B. is an employee of Verve Therapeutics and holds equity in Verve Therapeutics, Lyndra Therapeutics, Corner Therapeutics, and Cocoon Biotech. All other authors are employees of and hold equity in Verve Therapeutics. Verve Therapeutics has filed for patent protection related to various aspects of therapeutic base editing of ANGPTL3 and GalNAc-Lipid nanoparticle manufacture and delivery, including the patent publication WO2021178725.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

