DNA methylation markers for kidney function and progression of diabetic kidney disease
- PMID: 37188670
- PMCID: PMC10185566
- DOI: 10.1038/s41467-023-37837-7
DNA methylation markers for kidney function and progression of diabetic kidney disease
Abstract
Epigenetic markers are potential biomarkers for diabetes and related complications. Using a prospective cohort from the Hong Kong Diabetes Register, we perform two independent epigenome-wide association studies to identify methylation markers associated with baseline estimated glomerular filtration rate (eGFR) and subsequent decline in kidney function (eGFR slope), respectively, in 1,271 type 2 diabetes subjects. Here we show 40 (30 previously unidentified) and eight (all previously unidentified) CpG sites individually reach epigenome-wide significance for baseline eGFR and eGFR slope, respectively. We also develop a multisite analysis method, which selects 64 and 37 CpG sites for baseline eGFR and eGFR slope, respectively. These models are validated in an independent cohort of Native Americans with type 2 diabetes. Our identified CpG sites are near genes enriched for functional roles in kidney diseases, and some show association with renal damage. This study highlights the potential of methylation markers in risk stratification of kidney disease among type 2 diabetes individuals.
© 2023. The Author(s).
Conflict of interest statement
J.C.N.C. has received research grants and/or honoraria for consultancy and/or giving lectures from AstraZeneca, Bayer, Boehringer Ingelheim, Celltrion, Eli-Lilly, Hua Medicine, Lee Powder, Merck Serono, Merck Sharp & Dohme, Pfizer, Servier, Sanofi and Viatris, holds patents for using biomakers to predict risks of diabetes and its complications and is a co-founder of GemVCare, a biotechnology company partially supported by the Hong Kong Government startup fund. RCWM has received research grants for clinical trials from AstraZeneca, Bayer, MSD, Novo Nordisk, Sanofi, Tricida Inc., and honoraria for consultancy or lectures from AstraZeneca, Bayer, and Boehringer Ingelheim, all used to support diabetes research at the Chinese University of Hong Kong. RCWM is a co-founder of GemVCare, a technology start-up initiated with support from the Hong Kong Government Innovation and Technology Commission and its Technology Start-up Support Scheme for Universities (TSSSU). K.Y.L., J.C.N.C., K.Y.Y., and R.C.W.M. submitted a patent related to this study. The remaining authors declare no other competing interests.
Figures

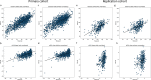

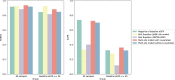
References
-
- Wheeler DC, et al. Effects of dapagliflozin on major adverse kidney and cardiovascular events in patients with diabetic and non-diabetic chronic kidney disease: a prespecified analysis from the DAPA-CKD trial. Lancet Diabetes Endocrinol. 2021;9:22–31. doi: 10.1016/S2213-8587(20)30369-7. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

