SIRT2 regulates extracellular vesicle-mediated liver-bone communication
- PMID: 37188819
- PMCID: PMC10229428
- DOI: 10.1038/s42255-023-00803-0
SIRT2 regulates extracellular vesicle-mediated liver-bone communication
Abstract
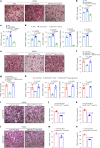
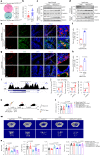
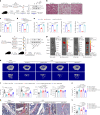
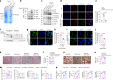
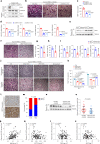
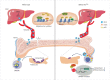
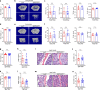
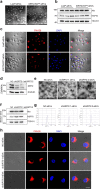
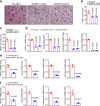
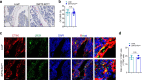
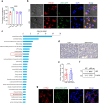
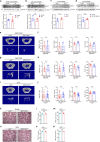
The interplay between liver and bone metabolism remains largely uncharacterized. Here, we uncover a mechanism of liver-bone crosstalk regulated by hepatocyte SIRT2. We demonstrate that hepatocyte SIRT2 expression is increased in aged mice and elderly humans. Liver-specific SIRT2 deficiency inhibits osteoclastogenesis and alleviates bone loss in mouse models of osteoporosis. We identify leucine-rich α-2-glycoprotein 1 (LRG1) as a functional cargo in hepatocyte-derived small extracellular vesicles (sEVs). In SIRT2-deficient hepatocytes, LRG1 levels in sEVs are upregulated, leading to increased transfer of LRG1 to bone-marrow-derived monocytes (BMDMs), and in turn, to inhibition of osteoclast differentiation via reduced nuclear translocation of NF-κB p65. Treatment with sEVs carrying high levels of LRG1 inhibits osteoclast differentiation in human BMDMs and in mice with osteoporosis, resulting in attenuated bone loss in mice. Furthermore, the plasma level of sEVs carrying LRG1 is positively correlated with bone mineral density in humans. Thus, drugs targeting hepatocyte-osteoclast communication may constitute a promising therapeutic strategy for primary osteoporosis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Comment in
-
Liver-bone crosstalk implicated in osteoporosis progression.Nat Rev Endocrinol. 2023 Aug;19(8):440. doi: 10.1038/s41574-023-00859-8. Nat Rev Endocrinol. 2023. PMID: 37258613 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

