FAIRness through automation: development of an automated medical data integration infrastructure for FAIR health data in a maximum care university hospital
- PMID: 37189148
- PMCID: PMC10186636
- DOI: 10.1186/s12911-023-02195-3
FAIRness through automation: development of an automated medical data integration infrastructure for FAIR health data in a maximum care university hospital
Abstract
Background: Secondary use of routine medical data is key to large-scale clinical and health services research. In a maximum care hospital, the volume of data generated exceeds the limits of big data on a daily basis. This so-called "real world data" are essential to complement knowledge and results from clinical trials. Furthermore, big data may help in establishing precision medicine. However, manual data extraction and annotation workflows to transfer routine data into research data would be complex and inefficient. Generally, best practices for managing research data focus on data output rather than the entire data journey from primary sources to analysis. To eventually make routinely collected data usable and available for research, many hurdles have to be overcome. In this work, we present the implementation of an automated framework for timely processing of clinical care data including free texts and genetic data (non-structured data) and centralized storage as Findable, Accessible, Interoperable, Reusable (FAIR) research data in a maximum care university hospital.
Methods: We identify data processing workflows necessary to operate a medical research data service unit in a maximum care hospital. We decompose structurally equal tasks into elementary sub-processes and propose a framework for general data processing. We base our processes on open-source software-components and, where necessary, custom-built generic tools.
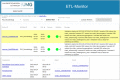
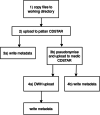
Results: We demonstrate the application of our proposed framework in practice by describing its use in our Medical Data Integration Center (MeDIC). Our microservices-based and fully open-source data processing automation framework incorporates a complete recording of data management and manipulation activities. The prototype implementation also includes a metadata schema for data provenance and a process validation concept. All requirements of a MeDIC are orchestrated within the proposed framework: Data input from many heterogeneous sources, pseudonymization and harmonization, integration in a data warehouse and finally possibilities for extraction or aggregation of data for research purposes according to data protection requirements.
Conclusion: Though the framework is not a panacea for bringing routine-based research data into compliance with FAIR principles, it provides a much-needed possibility to process data in a fully automated, traceable, and reproducible manner.
Keywords: Automated medical data processing; Electronic health record; Maximum care hospital; Medical data integration center; Medical data reuse; Medical informatics.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Traceable Research Data Sharing in a German Medical Data Integration Center With FAIR (Findability, Accessibility, Interoperability, and Reusability)-Geared Provenance Implementation: Proof-of-Concept Study.JMIR Form Res. 2023 Dec 7;7:e50027. doi: 10.2196/50027. JMIR Form Res. 2023. PMID: 38060305 Free PMC article.
-
[FAIR health data in the national and international data space].Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2024 Jun;67(6):710-720. doi: 10.1007/s00103-024-03884-8. Epub 2024 May 15. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2024. PMID: 38750239 Free PMC article. German.
-
Big Data Health Care Platform With Multisource Heterogeneous Data Integration and Massive High-Dimensional Data Governance for Large Hospitals: Design, Development, and Application.JMIR Med Inform. 2022 Apr 13;10(4):e36481. doi: 10.2196/36481. JMIR Med Inform. 2022. PMID: 35416792 Free PMC article.
-
INSPIRE datahub: a pan-African integrated suite of services for harmonising longitudinal population health data using OHDSI tools.Front Digit Health. 2024 Jan 29;6:1329630. doi: 10.3389/fdgth.2024.1329630. eCollection 2024. Front Digit Health. 2024. PMID: 38347885 Free PMC article. Review.
-
[A New Paradigm in Health Research: FAIR Data (Findable, Accessible, Interoperable, Reusable)].Acta Med Port. 2020 Dec 2;33(12):828-834. doi: 10.20344/amp.12910. Epub 2020 Dec 2. Acta Med Port. 2020. PMID: 33496252 Review. Portuguese.
Cited by
-
Research collaboration data platform ensuring general data protection.Sci Rep. 2024 May 24;14(1):11887. doi: 10.1038/s41598-024-61912-8. Sci Rep. 2024. PMID: 38789442 Free PMC article.
-
Proposal for Using AI to Assess Clinical Data Integrity and Generate Metadata: Algorithm Development and Validation.JMIR Med Inform. 2025 Jun 30;13:e60204. doi: 10.2196/60204. JMIR Med Inform. 2025. PMID: 40587839 Free PMC article.
-
Challenges and applications in generative AI for clinical tabular data in physiology.Pflugers Arch. 2025 Apr;477(4):531-542. doi: 10.1007/s00424-024-03024-w. Epub 2024 Oct 17. Pflugers Arch. 2025. PMID: 39417878 Free PMC article. Review.
-
Enhancing Clinical Data Infrastructure for AI Research: Comparative Evaluation of Data Management Architectures.J Med Internet Res. 2025 Aug 1;27:e74976. doi: 10.2196/74976. J Med Internet Res. 2025. PMID: 40749197 Free PMC article.
-
Integrating Clinical Data and Medical Imaging in Lung Cancer: Feasibility Study Using the Observational Medical Outcomes Partnership Common Data Model Extension.JMIR Med Inform. 2024 Jul 12;12:e59187. doi: 10.2196/59187. JMIR Med Inform. 2024. PMID: 38996330 Free PMC article.
References
-
- Cao Y, Jones C, Cuevas-Vicenttín V, Jones MB, Ludäscher B, McPhillips T, et al. DataONE: A Data Federation with Provenance Support. In: Mattoso M, Glavic B, et al., editors. Provenance and Annotation of Data and Processes IPAW 2016 Lecture Notes in Computer Science. Springer Cham; 2016. pp. 230–4.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

