Multivariate analysis and model building for classifying patients in the peroxisomal disorders X-linked adrenoleukodystrophy and Zellweger syndrome in Chinese pediatric patients
- PMID: 37189159
- PMCID: PMC10186734
- DOI: 10.1186/s13023-023-02673-x
Multivariate analysis and model building for classifying patients in the peroxisomal disorders X-linked adrenoleukodystrophy and Zellweger syndrome in Chinese pediatric patients
Erratum in
-
Correction: Multivariate analysis and model building for classifying patients in the peroxisomal disorders X-linked adrenoleukodystrophy and Zellweger syndrome in Chinese pediatric patients.Orphanet J Rare Dis. 2023 Jun 15;18(1):150. doi: 10.1186/s13023-023-02752-z. Orphanet J Rare Dis. 2023. PMID: 37322480 Free PMC article. No abstract available.
Abstract
Background: The peroxisome is a ubiquitous single membrane-enclosed organelle with an important metabolic role. Peroxisomal disorders represent a class of medical conditions caused by deficiencies in peroxisome function and are segmented into enzyme-and-transporter defects (defects in single peroxisomal proteins) and peroxisome biogenesis disorders (defects in the peroxin proteins, critical for normal peroxisome assembly and biogenesis). In this study, we employed multivariate supervised and non-supervised statistical methods and utilized mass spectrometry data of neurological patients, peroxisomal disorder patients (X-linked adrenoleukodystrophy and Zellweger syndrome), and healthy controls to analyze the role of common metabolites in peroxisomal disorders, to develop and refine a classification models of X-linked adrenoleukodystrophy and Zellweger syndrome, and to explore analytes with utility in rapid screening and diagnostics.
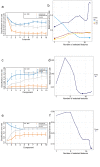
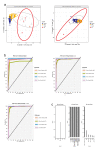
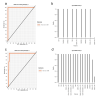
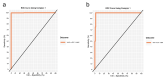
Results: T-SNE, PCA, and (sparse) PLS-DA, operated on mass spectrometry data of patients and healthy controls were utilized in this study. The performance of exploratory PLS-DA models was assessed to determine a suitable number of latent components and variables to retain for sparse PLS-DA models. Reduced-features (sparse) PLS-DA models achieved excellent classification performance of X-linked adrenoleukodystrophy and Zellweger syndrome patients.
Conclusions: Our study demonstrated metabolic differences between healthy controls, neurological patients, and peroxisomal disorder (X-linked adrenoleukodystrophy and Zellweger syndrome) patients, refined classification models and showed the potential utility of hexacosanoylcarnitine (C26:0-carnitine) as a screening analyte for Chinese patients in the context of a multivariate discriminant model predictive of peroxisomal disorders.
Keywords: C26: carnitine; Hexacosanoylcarnitine; Metabolomic signature; Newborn screening; PCA; PLS-DA; Sparse PLS-DA; Very long chain fatty acids; X-ALD; X-linked adrenoleukodystrophy; Zellweger syndrome; t-SNE.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

