The RUNX Family of Proteins, DNA Repair, and Cancer
- PMID: 37190015
- PMCID: PMC10136874
- DOI: 10.3390/cells12081106
The RUNX Family of Proteins, DNA Repair, and Cancer
Abstract
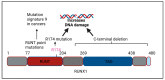
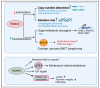
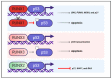
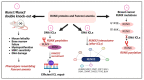
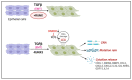
The RUNX family of transcription factors, including RUNX1, RUNX2, and RUNX3, are key regulators of development and can function as either tumor suppressors or oncogenes in cancer. Emerging evidence suggests that the dysregulation of RUNX genes can promote genomic instability in both leukemia and solid cancers by impairing DNA repair mechanisms. RUNX proteins control the cellular response to DNA damage by regulating the p53, Fanconi anemia, and oxidative stress repair pathways through transcriptional or non-transcriptional mechanisms. This review highlights the importance of RUNX-dependent DNA repair regulation in human cancers.
Keywords: DNA damage; DNA repair; Fanconi anemia; RUNX1; RUNX2; RUNX3; TGF-β; leukemia; p53; reactive oxygen species.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Novel biomarkers: the RUNX family as prognostic predictors in colorectal cancer.Front Immunol. 2024 Dec 9;15:1430136. doi: 10.3389/fimmu.2024.1430136. eCollection 2024. Front Immunol. 2024. PMID: 39822248 Free PMC article.
-
A Regulatory Role for RUNX1, RUNX3 in the Maintenance of Genomic Integrity.Adv Exp Med Biol. 2017;962:491-510. doi: 10.1007/978-981-10-3233-2_29. Adv Exp Med Biol. 2017. PMID: 28299675 Review.
-
RUNX Poly(ADP-Ribosyl)ation and BLM Interaction Facilitate the Fanconi Anemia Pathway of DNA Repair.Cell Rep. 2018 Aug 14;24(7):1747-1755. doi: 10.1016/j.celrep.2018.07.038. Cell Rep. 2018. PMID: 30110632
-
Role of RUNX Family Transcription Factors in DNA Damage Response.Mol Cells. 2020 Feb 29;43(2):99-106. doi: 10.14348/molcells.2019.0304. Mol Cells. 2020. PMID: 32024352 Free PMC article. Review.
-
Functional relationship between p53 and RUNX proteins.J Mol Cell Biol. 2019 Mar 1;11(3):224-230. doi: 10.1093/jmcb/mjy076. J Mol Cell Biol. 2019. PMID: 30535344 Free PMC article. Review.
Cited by
-
Evolutionary history of adenomas to colorectal cancer in FAP families.Front Genet. 2024 Jul 3;15:1391851. doi: 10.3389/fgene.2024.1391851. eCollection 2024. Front Genet. 2024. PMID: 39021676 Free PMC article.
-
Novel biomarkers: the RUNX family as prognostic predictors in colorectal cancer.Front Immunol. 2024 Dec 9;15:1430136. doi: 10.3389/fimmu.2024.1430136. eCollection 2024. Front Immunol. 2024. PMID: 39822248 Free PMC article.
-
Systematic decoding of functional enhancer connectomes and risk variants in human glioma.Nat Cell Biol. 2025 Aug 19. doi: 10.1038/s41556-025-01737-3. Online ahead of print. Nat Cell Biol. 2025. PMID: 40830415
-
RUNX2 as a novel biomarker for early identification of patients progressing to advanced-stage mycosis fungoides.Front Oncol. 2024 Oct 7;14:1421443. doi: 10.3389/fonc.2024.1421443. eCollection 2024. Front Oncol. 2024. PMID: 39435287 Free PMC article.
-
MicroRNA-150-3p enhances the antitumour effects of CGP57380 and is associated with a favourable prognosis in non-small cell lung cancer.Sci Rep. 2025 Jan 15;15(1):1973. doi: 10.1038/s41598-025-85793-7. Sci Rep. 2025. PMID: 39809860 Free PMC article.
References
-
- Tahirov T.H., Inoue-Bungo T., Morii H., Fujikawa A., Sasaki M., Kimura K., Shiina M., Sato K., Kumasaka T., Yamamoto M., et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell. 2001;104:755–767. doi: 10.1016/S0092-8674(01)00271-9. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

