Bioelectricity in Developmental Patterning and Size Control: Evidence and Genetically Encoded Tools in the Zebrafish Model
- PMID: 37190057
- PMCID: PMC10137051
- DOI: 10.3390/cells12081148
Bioelectricity in Developmental Patterning and Size Control: Evidence and Genetically Encoded Tools in the Zebrafish Model
Abstract
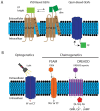
Developmental patterning is essential for regulating cellular events such as axial patterning, segmentation, tissue formation, and organ size determination during embryogenesis. Understanding the patterning mechanisms remains a central challenge and fundamental interest in developmental biology. Ion-channel-regulated bioelectric signals have emerged as a player of the patterning mechanism, which may interact with morphogens. Evidence from multiple model organisms reveals the roles of bioelectricity in embryonic development, regeneration, and cancers. The Zebrafish model is the second most used vertebrate model, next to the mouse model. The zebrafish model has great potential for elucidating the functions of bioelectricity due to many advantages such as external development, transparent early embryogenesis, and tractable genetics. Here, we review genetic evidence from zebrafish mutants with fin-size and pigment changes related to ion channels and bioelectricity. In addition, we review the cell membrane voltage reporting and chemogenetic tools that have already been used or have great potential to be implemented in zebrafish models. Finally, new perspectives and opportunities for bioelectricity research with zebrafish are discussed.
Keywords: GEVI; bioelectricity; chemogenetics; embryonic development; ion channels; long fin; optogenetics; patterning; pigment; short fin; zebrafish.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Bioelectricity in dental medicine: a narrative review.Biomed Eng Online. 2024 Jan 3;23(1):3. doi: 10.1186/s12938-023-01189-6. Biomed Eng Online. 2024. PMID: 38172866 Free PMC article. Review.
-
Potassium Channel-Associated Bioelectricity of the Dermomyotome Determines Fin Patterning in Zebrafish.Genetics. 2020 Aug;215(4):1067-1084. doi: 10.1534/genetics.120.303390. Epub 2020 Jun 16. Genetics. 2020. PMID: 32546498 Free PMC article.
-
Zebrafish Embryos Display Characteristic Bioelectric Signals during Early Development.Cells. 2022 Nov 12;11(22):3586. doi: 10.3390/cells11223586. Cells. 2022. PMID: 36429015 Free PMC article.
-
Visualization of Cellular Electrical Activity in Zebrafish Early Embryos and Tumors.J Vis Exp. 2018 Apr 25;(134):57330. doi: 10.3791/57330. J Vis Exp. 2018. PMID: 29757272 Free PMC article.
-
Molecular bioelectricity in developmental biology: new tools and recent discoveries: control of cell behavior and pattern formation by transmembrane potential gradients.Bioessays. 2012 Mar;34(3):205-17. doi: 10.1002/bies.201100136. Epub 2012 Jan 11. Bioessays. 2012. PMID: 22237730 Free PMC article. Review.
Cited by
-
From the Microbiome to the Electrome: Implications for the Microbiota-Gut-Brain Axis.Int J Mol Sci. 2024 Jun 5;25(11):6233. doi: 10.3390/ijms25116233. Int J Mol Sci. 2024. PMID: 38892419 Free PMC article.
-
Bioelectricity in dental medicine: a narrative review.Biomed Eng Online. 2024 Jan 3;23(1):3. doi: 10.1186/s12938-023-01189-6. Biomed Eng Online. 2024. PMID: 38172866 Free PMC article. Review.
-
Evolution of two-pore domain potassium channels and their gene expression in zebrafish embryos.Dev Dyn. 2024 Aug;253(8):722-749. doi: 10.1002/dvdy.690. Epub 2024 Jan 25. Dev Dyn. 2024. PMID: 38270285 Free PMC article.
-
Bioelectricity is a universal multifaced signaling cue in living organisms.Mol Biol Cell. 2025 Feb 1;36(2):pe2. doi: 10.1091/mbc.E23-08-0312. Mol Biol Cell. 2025. PMID: 39873662 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

