IKKβ Inhibition Attenuates Epithelial Mesenchymal Transition of Human Stem Cell-Derived Retinal Pigment Epithelium
- PMID: 37190063
- PMCID: PMC10136838
- DOI: 10.3390/cells12081155
IKKβ Inhibition Attenuates Epithelial Mesenchymal Transition of Human Stem Cell-Derived Retinal Pigment Epithelium
Abstract
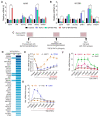
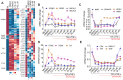
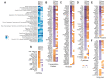
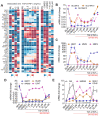
Epithelial-mesenchymal transition (EMT), which is well known for its role in embryonic development, malignant transformation, and tumor progression, has also been implicated in a variety of retinal diseases, including proliferative vitreoretinopathy (PVR), age-related macular degeneration (AMD), and diabetic retinopathy. EMT of the retinal pigment epithelium (RPE), although important in the pathogenesis of these retinal conditions, is not well understood at the molecular level. We and others have shown that a variety of molecules, including the co-treatment of human stem cell-derived RPE monolayer cultures with transforming growth factor beta (TGF-β) and the inflammatory cytokine tumor necrosis factor alpha (TNF-α), can induce RPE-EMT; however, small molecule inhibitors of RPE-EMT have been less well studied. Here, we demonstrate that BAY651942, a small molecule inhibitor of nuclear factor kapa-B kinase subunit beta (IKKβ) that selectively targets NF-κB signaling, can modulate TGF-β/TNF-α-induced RPE-EMT. Next, we performed RNA-seq studies on BAY651942 treated hRPE monolayers to dissect altered biological pathways and signaling events. Further, we validated the effect of IKKβ inhibition on RPE-EMT-associated factors using a second IKKβ inhibitor, BMS345541, with RPE monolayers derived from an independent stem cell line. Our data highlights the fact that pharmacological inhibition of RPE-EMT restores RPE identity and may provide a promising approach for treating retinal diseases that involve RPE dedifferentiation and EMT.
Keywords: PVR and AMD; TGF–β/–α; differentiation; epithelial-mesenchymal transition; kinase inhibitors; retinal pigment epithelium; stem cells; transcriptomics.
Conflict of interest statement
RCT is an employee of Caris Life Sciences.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

