Myeloid NGS Analyses of Paired Samples from Bone Marrow and Peripheral Blood Yield Concordant Results: A Prospective Cohort Analysis of the AGMT Study Group
- PMID: 37190237
- PMCID: PMC10136651
- DOI: 10.3390/cancers15082305
Myeloid NGS Analyses of Paired Samples from Bone Marrow and Peripheral Blood Yield Concordant Results: A Prospective Cohort Analysis of the AGMT Study Group
Abstract
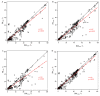
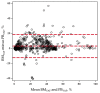
Background: Next generation sequencing (NGS) has become indispensable for diagnosis, risk stratification, prognostication, and monitoring of response in patients with myeloid neoplasias. Guidelines require bone marrow evaluations for the above, which are often not performed outside of clinical trials, indicating a need for surrogate samples. Methods: Myeloid NGS analyses (40 genes and 29 fusion drivers) of 240 consecutive, non-selected, prospectively collected, paired bone marrow/peripheral blood samples were compared. Findings: Very strong correlation (r = 0.91, p < 0.0001), high concordance (99.6%), sensitivity (98.8%), specificity (99.9%), positive predictive value (99.8%), and negative predictive value (99.6%) between NGS analyses of paired samples was observed. A total of 9/1321 (0.68%) detected mutations were discordant, 8 of which had a variant allele frequency (VAF) ≤ 3.7%. VAFs between peripheral blood and bone marrow samples were very strongly correlated in the total cohort (r = 0.93, p = 0.0001) and in subgroups without circulating blasts (r = 0.92, p < 0.0001) or with neutropenia (r = 0.88, p < 0.0001). There was a weak correlation between the VAF of a detected mutation and the blast count in either the peripheral blood (r = 0.19) or the bone marrow (r = 0.11). Interpretation: Peripheral blood samples can be used to molecularly classify and monitor myeloid neoplasms via NGS without loss of sensitivity/specificity, even in the absence of circulating blasts or in neutropenic patients.
Keywords: acute myeloid leukemia (AML); bone marrow; concordance; diagnosis; myelodysplastic syndromes/neoplasms (MDS); myeloid neoplasias; next generation sequencing (NGS); peripheral blood; prognosis.
Conflict of interest statement
B.J.-G.: No potential conflict of interest; M.L.: Honoraria from BMS, Celgene, Gilead, Takeda and Novartis; Travel support: Celgene and Novartis; F.J.G.: No potential conflict of interest; N.Z.: No potential conflict of interest; T.D.: No potential conflict of interest; S.H.: No potential conflict of interest; A.R.: No potential conflict of interest; T.M.: Honoraria from AbbVie and Celgene/BMS; A.E.: Honoraria, consultancy, and travel support from AbbVie and BMS/Celgene; M.D.: No potential conflict of interest; J.L.-S.: No potential conflict of interest; R.G.: Honoraria from AbbVie, Amgen, AstraZeneca, BMS/Celgene, Daiichi Sankyo, Gilead, Merck, Novartis, Roche, Takeda, BMS, MSD, Sandoz, Gilead; Research funding from Celgene, Roche, Merck, Novartis, MSD, Sandoz, and Takeda; Consulting: AbbVie, AstraZeneca, BMS/Celgene, Novartis, Roche, Takeda, Janssen, MSD, Merck, Gilead, Daiichi Sankyo; Travel support from AbbVie, Amgen, AstraZeneca, BMS/Celgene, Daiichi Sankyo, Gilead, Janssen Cilag, MSD, Novartis, and Roche; L.P.: Honoraria from AbbVie, BMS, and Novartis.
Figures




References
-
- Swerdlow S.H., Campo E., Pileri S.A., Harris N.L., Stein H., Siebert R., Advani R., Ghielmini M., Salles G.A., Zelenetz A.D., et al. The 2016 Revision of the World Health Organization Classification of Lymphoid Neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. - DOI - PMC - PubMed
-
- Pleyer L., Leisch M., Kourakli A., Padron E., Maciejewski J.P., Xicoy Cirici B., Kaivers J., Ungerstedt J., Heibl S., Patiou P., et al. Outcomes of Patients with Chronic Myelomonocytic Leukaemia Treated with Non-Curative Therapies: A Retrospective Cohort Study. Lancet Haematol. 2021;8:e135–e148. doi: 10.1016/S2352-3026(20)30374-4. - DOI - PubMed
-
- Pleyer L., Neureiter D., Faber V., Greil R. Myelodysplastic Syndromes (MDS) In: Greil R., Pleyer L., Faber V., Neureiter D., editors. Chronic Myeloid Neoplasias and Clonal Overlap Syndromes. Springer; Vienna, Austria: 2010. pp. 153–222.
-
- Valent P., Orazi A., Savona M.R., Patnaik M.M., Onida F., van de Loosdrecht A.A., Haase D., Haferlach T., Elena C., Pleyer L., et al. Proposed Diagnostic Criteria for Classical Chronic Myelomonocytic Leukemia (CMML), CMML Variants and Pre-CMML Conditions. Haematologica. 2019;104:1935–1949. doi: 10.3324/haematol.2019.222059. - DOI - PMC - PubMed
-
- Pleyer L., Döhner H., Dombret H., Seymour J., Schuh A., Beach C., Swern A., Burgstaller S., Stauder R., Girschikofsky M., et al. Azacitidine for Front-Line Therapy of Patients with AML: Reproducible Efficacy Established by Direct Comparison of International Phase 3 Trial Data with Registry Data from the Austrian Azacitidine Registry of the AGMT Study Group. IJMS. 2017;18:415. doi: 10.3390/ijms18020415. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

