Biomarkers of Tumor Heterogeneity in Glioblastoma Multiforme Cohort of TCGA
- PMID: 37190318
- PMCID: PMC10137245
- DOI: 10.3390/cancers15082387
Biomarkers of Tumor Heterogeneity in Glioblastoma Multiforme Cohort of TCGA
Abstract
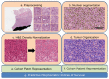
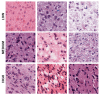
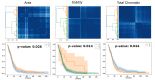
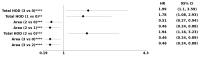
Tumor Whole Slide Images (WSI) are often heterogeneous, which hinders the discovery of biomarkers in the presence of confounding clinical factors. In this study, we present a pipeline for identifying biomarkers from the Glioblastoma Multiforme (GBM) cohort of WSIs from TCGA archive. The GBM cohort endures many technical artifacts while the discovery of GBM biomarkers is challenged because "age" is the single most confounding factor for predicting outcomes. The proposed approach relies on interpretable features (e.g., nuclear morphometric indices), effective similarity metrics for heterogeneity analysis, and robust statistics for identifying biomarkers. The pipeline first removes artifacts (e.g., pen marks) and partitions each WSI into patches for nuclear segmentation via an extended U-Net for subsequent quantitative representation. Given the variations in fixation and staining that can artificially modulate hematoxylin optical density (HOD), we extended Navab's Lab method to normalize images and reduce the impact of batch effects. The heterogeneity of each WSI is then represented either as probability density functions (PDF) per patient or as the composition of a dictionary predicted from the entire cohort of WSIs. For PDF- or dictionary-based methods, morphometric subtypes are constructed based on distances computed from optimal transport and linkage analysis or consensus clustering with Euclidean distances, respectively. For each inferred subtype, Kaplan-Meier and/or the Cox regression model are used to regress the survival time. Since age is the single most important confounder for predicting survival in GBM and there is an observed violation of the proportionality assumption in the Cox model, we use both age and age-squared coupled with the Likelihood ratio test and forest plots for evaluating competing statistics. Next, the PDF- and dictionary-based methods are combined to identify biomarkers that are predictive of survival. The combined model has the advantage of integrating global (e.g., cohort scale) and local (e.g., patient scale) attributes of morphometric heterogeneity, coupled with robust statistics, to reveal stable biomarkers. The results indicate that, after normalization of the GBM cohort, mean HOD, eccentricity, and cellularity are predictive of survival. Finally, we also stratified the GBM cohort as a function of EGFR expression and published genomic subtypes to reveal genomic-dependent morphometric biomarkers.
Keywords: Glioblastoma Multiforme; TCGA; biomarker; tumor heterogeneity; whole slide imaging.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. - DOI - PMC - PubMed
-
- Zhu X., Yao J., Huang J. Deep convolutional neural network for survival analysis with pathological images; Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); Shenzhen, China. 15–18 December 2016; Piscataway, NJ, USA: IEEE; 2016. pp. 544–547.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

