Structural Insights into the Assembly of the African Swine Fever Virus Inner Capsid
- PMID: 37191520
- PMCID: PMC10308890
- DOI: 10.1128/jvi.00268-23
Structural Insights into the Assembly of the African Swine Fever Virus Inner Capsid
Abstract
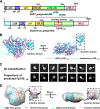
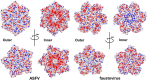
African swine fever virus (ASFV), the cause of a highly contagious hemorrhagic and fatal disease of domestic pigs, has a complex multilayer structure. The inner capsid of ASFV located underneath the inner membrane enwraps the genome-containing nucleoid and is likely the assembly of proteolytic products from the virally encoded polyproteins pp220 and pp62. Here, we report the crystal structure of ASFV p150△NC, a major middle fragment of the pp220 proteolytic product p150. The structure of ASFV p150△NC contains mainly helices and has a triangular plate-like shape. The triangular plate is approximately 38 Å in thickness, and the edge of the triangular plate is approximately 90 Å long. The structure of ASFV p150△NC is not homologous to any of the known viral capsid proteins. Further analysis of the cryo-electron microscopy maps of the ASFV and the homologous faustovirus inner capsids revealed that p150 or the p150-like protein of faustovirus assembles to form screwed propeller-shaped hexametric and pentametric capsomeres of the icosahedral inner capsids. Complexes of the C terminus of p150 and other proteolytic products of pp220 likely mediate interactions between the capsomeres. Together, these findings provide new insights into the assembling of ASFV inner capsid and provide a reference for understanding the assembly of the inner capsids of nucleocytoplasmic large DNA viruses (NCLDV). IMPORTANCE African swine fever virus has caused catastrophic destruction to the pork industry worldwide since it was first discovered in Kenya in 1921. The architecture of ASFV is complicated, with two protein shells and two membrane envelopes. Currently, mechanisms involved in the assembly of the ASFV inner core shell are less understood. The structural studies of the ASFV inner capsid protein p150 performed in this research enable the building of a partial model of the icosahedral ASFV inner capsid, which provides a structural basis for understanding the structure and assembly of this complex virion. Furthermore, the structure of ASFV p150△NC represents a new type of fold for viral capsid assembly, which could be a common fold for the inner capsid assembly of nucleocytoplasmic large DNA viruses (NCLDV) and would facilitate the development of vaccine and antivirus drugs against these complex viruses.
Keywords: African swine fever virus; cryogenic electron microscopy; inner capsid assembly; new viral capsid scaffold; nucleocytoplasmic large DNA viruses; virion structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
The cryo-EM structure of African swine fever virus unravels a unique architecture comprising two icosahedral protein capsids and two lipoprotein membranes.J Biol Chem. 2020 Jan 3;295(1):1-12. doi: 10.1074/jbc.AC119.011196. Epub 2019 Oct 24. J Biol Chem. 2020. PMID: 31649031 Free PMC article.
-
African swine fever virus polyproteins pp220 and pp62 assemble into the core shell.J Virol. 2002 Dec;76(24):12473-82. doi: 10.1128/jvi.76.24.12473-12482.2002. J Virol. 2002. PMID: 12438573 Free PMC article.
-
African swine fever virus A137R assembles into a dodecahedron cage.J Virol. 2024 Mar 19;98(3):e0153623. doi: 10.1128/jvi.01536-23. Epub 2024 Feb 5. J Virol. 2024. PMID: 38315014 Free PMC article.
-
Structures and Functional Diversities of ASFV Proteins.Viruses. 2021 Oct 21;13(11):2124. doi: 10.3390/v13112124. Viruses. 2021. PMID: 34834930 Free PMC article. Review.
-
African swine fever virus morphogenesis.Virus Res. 2013 Apr;173(1):29-41. doi: 10.1016/j.virusres.2012.09.016. Epub 2012 Oct 8. Virus Res. 2013. PMID: 23059353 Review.
Cited by
-
African Swine Fever Virus Immunosuppression and Virulence-Related Gene.Curr Issues Mol Biol. 2024 Jul 31;46(8):8268-8281. doi: 10.3390/cimb46080488. Curr Issues Mol Biol. 2024. PMID: 39194705 Free PMC article. Review.
-
Comprehensive Characterization of the Genetic Landscape of African Swine Fever Virus: Insights into Infection Dynamics, Immunomodulation, Virulence and Genes with Unknown Function.Animals (Basel). 2024 Jul 26;14(15):2187. doi: 10.3390/ani14152187. Animals (Basel). 2024. PMID: 39123713 Free PMC article. Review.
-
A Pool of Bacterium-like Particles Displaying African Swine Fever Virus Antigens Induces Both Humoral and Cellular Immune Responses in Pigs.Vaccines (Basel). 2024 Dec 24;13(1):5. doi: 10.3390/vaccines13010005. Vaccines (Basel). 2024. PMID: 39852784 Free PMC article.
-
Genetic Variations of African Swine Fever Virus: Major Challenges and Prospects.Viruses. 2024 Jun 4;16(6):913. doi: 10.3390/v16060913. Viruses. 2024. PMID: 38932205 Free PMC article. Review.
-
Advancement in the development of gene/protein-based vaccines against African swine fever virus.Curr Res Microb Sci. 2024 Mar 12;6:100232. doi: 10.1016/j.crmicr.2024.100232. eCollection 2024. Curr Res Microb Sci. 2024. PMID: 38545259 Free PMC article. Review.
References
-
- Eustace Montgomery R. 1921. On a form of swine fever occurring in British East Africa (Kenya Colony). J Comp Pathol Ther 34:159–191. doi:10.1016/S0368-1742(21)80031-4. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

