Metabolic adaptation to tyrosine kinase inhibition in leukemia stem cells
- PMID: 37192295
- PMCID: PMC10447615
- DOI: 10.1182/blood.2022018196
Metabolic adaptation to tyrosine kinase inhibition in leukemia stem cells
Abstract
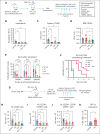
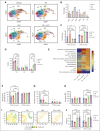
Tyrosine kinase inhibitors (TKIs) are very effective in treating chronic myelogenous leukemia (CML), but primitive, quiescent leukemia stem cells persist as a barrier to the cure. We performed a comprehensive evaluation of metabolic adaptation to TKI treatment and its role in CML hematopoietic stem and progenitor cell persistence. Using a CML mouse model, we found that glycolysis, glutaminolysis, the tricarboxylic acid cycle, and oxidative phosphorylation (OXPHOS) were initially inhibited by TKI treatment in CML-committed progenitors but were restored with continued treatment, reflecting both selection and metabolic reprogramming of specific subpopulations. TKI treatment selectively enriched primitive CML stem cells with reduced metabolic gene expression. Persistent CML stem cells also showed metabolic adaptation to TKI treatment through altered substrate use and mitochondrial respiration maintenance. Evaluation of transcription factors underlying these changes helped detect increased HIF-1 protein levels and activity in TKI-treated stem cells. Treatment with an HIF-1 inhibitor in combination with TKI treatment depleted murine and human CML stem cells. HIF-1 inhibition increased mitochondrial activity and reactive oxygen species (ROS) levels, reduced quiescence, increased cycling, and reduced the self-renewal and regenerating potential of dormant CML stem cells. We, therefore, identified the HIF-1-mediated inhibition of OXPHOS and ROS and maintenance of CML stem cell dormancy and repopulating potential as a key mechanism of CML stem cell adaptation to TKI treatment. Our results identify a key metabolic dependency in CML stem cells persisting after TKI treatment that can be targeted to enhance their elimination.
© 2023 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures






Comment in
-
Food for thought (and CML survival).Blood. 2023 Aug 10;142(6):502-504. doi: 10.1182/blood.2023021051. Blood. 2023. PMID: 37561542 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

