Inhibition of histone methyltransferase SETD8 represses DNA virus replication
- PMID: 37193046
- PMCID: PMC10120311
- DOI: 10.1016/j.cellin.2022.100033
Inhibition of histone methyltransferase SETD8 represses DNA virus replication
Erratum in
-
Erratum regarding missing declaration of interests in previously published articles.Cell Insight. 2024 Jan 30;3(1):100148. doi: 10.1016/j.cellin.2024.100148. eCollection 2024 Feb. Cell Insight. 2024. PMID: 38323319 Free PMC article.
Abstract
Multiple diseases, such as cancer and neural degeneration diseases, are related with the latent infection of DNA viruses. However, it is still difficult to clean up the latent DNA viruses and new anti-viral strategies are critical for disease treatment. Here, we screen a pool of small chemical molecules and identify UNC0379, an inhibitor for histone H4K20 methyltransferase SETD8, as an effective inhibitor for multiple DNA viruses. UNC0379 not only enhances the expression of anti-viral genes in THP-1 cells, but also repress DNA virus replication in multiple cell lines with defects in cGAS pathway. We prove that SETD8 promotes DNA virus replication in a manner dependent on its enzyme activity. Our results further indicated that SETD8 is required for PCNA stability, one factor critical for viral DNA replication. Viral infection stimulates the interaction between SETD8 and PCNA and thus enhances PCNA stability and viral DNA replication. Taken together, our study reveals a new mechanism for regulating viral DNA replication and provides a potential strategy for treatment of diseases related with DNA viruses.
Keywords: DNA replication; DNA virus; PCNA; SETD8; UNC0379.
© 2022 The Author(s).
Figures


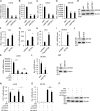
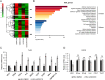

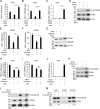

References
-
- Cai X., Chiu Y.H., Chen Z.J. The cgas-cgamp-Sting pathway of cytosolic Dna sensing and signaling. Mol. Cell. 2014;54:289–296. - PubMed
-
- Daigle S.R., Olhava E.J., Therkelsen C.A., Basavapathruni A., Jin L., Boriack-Sjodin P.A., Allain C.J., Klaus C.R., Raimondi A., Scott M.P., Waters N.J., Chesworth R., Moyer M.P., Copeland R.A., Richon V.M., Pollock R.M. Potent inhibition of Dot1l as treatment of Mll-fusion leukemia. Blood. 2013;122:1017–1025. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

