Emerging SARS-CoV-2 variants: Why, how, and what's next?
- PMID: 37193049
- PMCID: PMC9057926
- DOI: 10.1016/j.cellin.2022.100029
Emerging SARS-CoV-2 variants: Why, how, and what's next?
Erratum in
-
Erratum regarding missing declaration of interests in previously published articles.Cell Insight. 2024 Jan 30;3(1):100148. doi: 10.1016/j.cellin.2024.100148. eCollection 2024 Feb. Cell Insight. 2024. PMID: 38323319 Free PMC article.
-
Corrigendum to previous published articles.Cell Insight. 2025 Jan 11;4(2):100225. doi: 10.1016/j.cellin.2024.100225. eCollection 2025 Apr. Cell Insight. 2025. PMID: 39881711 Free PMC article.
Abstract
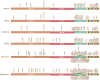
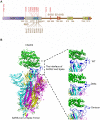
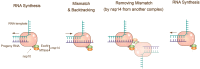
The emergence of the SARS-CoV-2 Omicron variant poses a striking threat to human society. More than 30 mutations in the Spike protein of the Omicron variant severely compromised the protective immunity elicited by either vaccination or prior infection. The persistent viral evolutionary trajectory generates Omicron-associated lineages, such as BA.1 and BA.2. Moreover, the virus recombination upon Delta and Omicron co-infections has been reported lately, although the impact remains to be assessed. This minireview summarizes the characteristics, evolution and mutation control, and immune evasion mechanisms of SARS-CoV-2 variants, which will be helpful for the in-depth understanding of the SARS-CoV-2 variants and policy-making related to COVID-19 pandemic control.
Keywords: Deltacron; Mutation rate; Mutations in the Spike; Omicron; SARS-CoV-2 variants.
© 2022 Published by Elsevier B.V. on behalf of Wuhan University.
Figures
References
-
- Abdullah F., et al. International Journal of Infectious Diseases : IJID : Official Publication of the International Society for Infectious Diseases. vol. 116. 2022. Decreased severity of disease during the first global omicron variant covid-19 outbreak in a large hospital in Tshwane, South Africa; pp. 38–42. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

