Endomembrane remodeling in SARS-CoV-2 infection
- PMID: 37193051
- PMCID: PMC9112566
- DOI: 10.1016/j.cellin.2022.100031
Endomembrane remodeling in SARS-CoV-2 infection
Erratum in
-
Erratum regarding missing declaration of interests in previously published articles.Cell Insight. 2024 Jan 30;3(1):100148. doi: 10.1016/j.cellin.2024.100148. eCollection 2024 Feb. Cell Insight. 2024. PMID: 38323319 Free PMC article.
Abstract
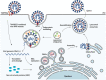
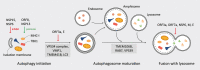
During severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, the viral proteins intimately interact with host factors to remodel the endomembrane system at various steps of the viral lifecycle. The entry of SARS-CoV-2 can be mediated by endocytosis-mediated internalization. Virus-containing endosomes then fuse with lysosomes, in which the viral S protein is cleaved to trigger membrane fusion. Double-membrane vesicles generated from the ER serve as platforms for viral replication and transcription. Virions are assembled at the ER-Golgi intermediate compartment and released through the secretory pathway and/or lysosome-mediated exocytosis. In this review, we will focus on how SARS-CoV-2 viral proteins collaborate with host factors to remodel the endomembrane system for viral entry, replication, assembly and egress. We will also describe how viral proteins hijack the host cell surveillance system-the autophagic degradation pathway-to evade destruction and benefit virus production. Finally, potential antiviral therapies targeting the host cell endomembrane system will be discussed.
Keywords: Autophagy; Coronavirus; DMV; Endocytosis; SARS-CoV-2.
© 2022 The Author(s).
Figures


References
-
- Baggen J., Persoons L., Vanstreels E., Jansen S., Van Looveren D., Boeckx B., Geudens V., De Man J., Jochmans D., Wauters J., Wauters E., Vanaudenaerde B.M., Lambrechts D., Neyts J., Dallmeier K., Thibaut H.J., Jacquemyn M., Maes P., Daelemans D. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat. Genet. 2021;53:435–444. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

