Cas12a-based one-pot SNP detection with high accuracy
- PMID: 37193068
- PMCID: PMC10134196
- DOI: 10.1016/j.cellin.2023.100080
Cas12a-based one-pot SNP detection with high accuracy
Erratum in
-
Corrigendum to previous published articles.Cell Insight. 2025 Jan 11;4(2):100225. doi: 10.1016/j.cellin.2024.100225. eCollection 2025 Apr. Cell Insight. 2025. PMID: 39881711 Free PMC article.
Abstract
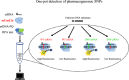
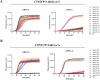
CRISPR-Cas12a based one-pot detection system has been used in nucleic acid detection and diagnosis. However, it is not sensitive enough to distinguish single nucleotide polymorphisms (SNP), which has greatly restricted its application. To overcome these limitations, we engineered a LbCas12a variant with enhanced sensitivity against SNP, named seCas12a (sensitive Cas12a). SeCas12a-based one-pot SNP detection system is a versatile platform that could use both canonical and non-canonical PAM, and was almost not limited by mutation types to distinguish SNPs located between position 1 to 17. The use of truncated crRNA further improved SNP specificity of seCas12a. Mechanistically, we found only when the cis-cleavage was at low level between 0.01min-1 and 0.0006 min-1, a good signal-to-noise ratio can be achieved in one-pot test. SeCas12a-based one-pot SNP detection system was applied to detect pharmacogenomic SNPs in human clinical samples. Of thirteen donors tested in two different SNPs, the seCas12a mediated one-pot system could faithfully detect the SNPs in 30 min with 100% accuracy.
© 2023 The Authors.
Conflict of interest statement
Y.Z., H.Y. and H.-X.Z. have a filed patent application on seCas12a-based SNP detection through Wuhan University (Patent No. 202211514174.0). The other authors declare no competing interests.
Figures




References
-
- Ablin J., Cabili S., Lagziel A., Peretz H. Warfarin therapy in a patient homozygous for the CYP2C9∗3 allele. The Israel Medical Association Journal. 2002;4:139–141. - PubMed
-
- Furuya H., FernandezSalguero P., Gregory W., Taber H., Steward A., Gonzalez F.J., Idle J.R. Genetic polymorphism of CYP2C9 and its effect on warfarin maintenance dose requirement in patients undergoing anticoagulation therapy. Pharmacogenetics. 1995;5:389–392. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

