The deubiquitinase OTUD4 inhibits the expression of antimicrobial peptides in Paneth cells to support intestinal inflammation and bacterial infection
- PMID: 37193092
- PMCID: PMC10123543
- DOI: 10.1016/j.cellin.2023.100100
The deubiquitinase OTUD4 inhibits the expression of antimicrobial peptides in Paneth cells to support intestinal inflammation and bacterial infection
Erratum in
-
Corrigendum to previous published articles.Cell Insight. 2025 Jan 11;4(2):100225. doi: 10.1016/j.cellin.2024.100225. eCollection 2025 Apr. Cell Insight. 2025. PMID: 39881711 Free PMC article.
Abstract
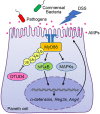
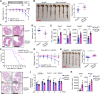
Dysfunction of the intestinal epithelial barrier causes microbial invasion that would lead to inflammation in the gut. Antimicrobial peptides (AMPs) are essential components of the intestinal epithelial barrier, while the regulatory mechanisms of AMPs expression are not fully characterized. Here, we report that the ovarian tumor family deubiquitinase 4 (OTUD4) in Paneth cells restricts the expression of AMPs and thereby promotes experimental colitis and bacterial infection. OTUD4 is upregulated in the inflamed mucosa of ulcerative colitis patients and in the colon of mice treated with dextran sulfate sodium salt (DSS). Knockout of OTUD4 promotes the expression of AMPs in intestinal organoids after stimulation with lipopolysaccharide (LPS) or peptidoglycan (PGN) and in the intestinal epithelial cells (IECs) of mice after DSS treatment or Salmonella typhimurium (S.t.) infection. Consistently, Vil-Cre;Otud4fl/fl mice and Def-Cre;Otud4fl/fl mice exhibit hyper-resistance to DSS-induced colitis and S.t. infection compared to Otud4fl/fl mice. Mechanistically, knockout of OTUD4 results in hyper K63-linked ubiquitination of MyD88 and increases the activation of NF-κB and MAPKs to promote the expression of AMPs. These findings collectively highlight an indispensable role of OTUD4 in Paneth cells to modulate AMPs production and indicate OTUD4 as a potential target for gastrointestinal inflammation and bacterial infection.
Keywords: Antimicrobial peptides; Bacterial infection; Colitis; Deubiquitination; MyD88; OTUD4.
© 2023 The Authors.
Figures




References
-
- Annese V. Genetics and epigenetics of IBD. Pharmacological Research. 2020;159 - PubMed
-
- Caruso R., Lo B.C., Nunez G. Host-microbiota interactions in inflammatory bowel disease. Nature Reviews Immunology. 2020;20:411–426. - PubMed
-
- Chen J., Zhai Z., Long H., Yang G., Deng B., Deng J. Inducible expression of defensins and cathelicidins by nutrients and associated regulatory mechanisms. Peptides. 2020;123 - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

