Trim25 restricts rabies virus replication by destabilizing phosphoprotein
- PMID: 37193556
- PMCID: PMC10120326
- DOI: 10.1016/j.cellin.2022.100057
Trim25 restricts rabies virus replication by destabilizing phosphoprotein
Erratum in
-
Erratum regarding missing declaration of interests in previously published articles.Cell Insight. 2024 Jan 30;3(1):100148. doi: 10.1016/j.cellin.2024.100148. eCollection 2024 Feb. Cell Insight. 2024. PMID: 38323319 Free PMC article.
Abstract
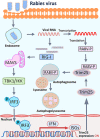
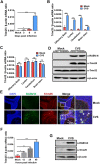
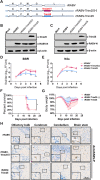
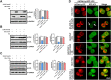
Tripartite motif-containing protein 25 (Trim25) is an E3 ubiquitin ligase that activates retinoid acid-inducible gene I (RIG-I) and promotes the antiviral interferon response. Recent studies have shown that Trim25 can bind and degrade viral proteins, suggesting a different mechanism of Trim25 on its antiviral effects. In this study, Trim25 expression was upregulated in cells and mouse brains after rabies virus (RABV) infection. Moreover, expression of Trim25 limited RABV replication in cultured cells. Overexpression of Trim25 caused attenuated viral pathogenicity in a mouse model that was intramuscularly injected with RABV. Further experiments confirmed that Trim25 inhibited RABV replication via two different mechanisms: an E3 ubiquitin ligase-dependent mechanism and an E3 ubiquitin ligase-independent mechanism. Specifically, the CCD domain of Trim25 interacted with RABV phosphoprotein (RABV-P) at amino acid (AA) position at 72 and impaired the stability of RABV-P via complete autophagy. This study reveals a novel mechanism by which Trim25 restricts RABV replication by destabilizing RABV-P, which is independent of its E3 ubiquitin ligase activity.
Keywords: Autophagy; Phosphoprotein; Protein stability; Rabies virus; Trim25.
© 2022 The Authors.
Figures





References
-
- Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M., et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. - PubMed
LinkOut - more resources
Full Text Sources

