Molecular signatures of post-traumatic stress disorder in war-zone-exposed veteran and active-duty soldiers
- PMID: 37196634
- PMCID: PMC10213980
- DOI: 10.1016/j.xcrm.2023.101045
Molecular signatures of post-traumatic stress disorder in war-zone-exposed veteran and active-duty soldiers
Abstract
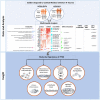
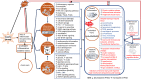
Post-traumatic stress disorder (PTSD) is a multisystem syndrome. Integration of systems-level multi-modal datasets can provide a molecular understanding of PTSD. Proteomic, metabolomic, and epigenomic assays are conducted on blood samples of two cohorts of well-characterized PTSD cases and controls: 340 veterans and 180 active-duty soldiers. All participants had been deployed to Iraq and/or Afghanistan and exposed to military-service-related criterion A trauma. Molecular signatures are identified from a discovery cohort of 218 veterans (109/109 PTSD+/-). Identified molecular signatures are tested in 122 separate veterans (62/60 PTSD+/-) and in 180 active-duty soldiers (PTSD+/-). Molecular profiles are computationally integrated with upstream regulators (genetic/methylation/microRNAs) and functional units (mRNAs/proteins/metabolites). Reproducible molecular features of PTSD are identified, including activated inflammation, oxidative stress, metabolic dysregulation, and impaired angiogenesis. These processes may play a role in psychiatric and physical comorbidities, including impaired repair/wound healing mechanisms and cardiovascular, metabolic, and psychiatric diseases.
Keywords: active duty; angiogenesis; inflammatory response; metabolic dysregulation; molecular signature; multi-omics; oxidative stress; post-traumatic stress disorder; veterans; wound healing.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests
Figures




References
-
- PTSD N.C.f. How common is PTSD? 2022. https://www.ptsd.va.gov/understand/common/common_veterans.asp
-
- Somvanshi P.R., Mellon S.H., Flory J.D., Abu-Amara D., PTSD Systems Biology Consortium; Wolkowitz O.M., Yehuda R., Jett M., Hood L., Marmar C., Doyle F.J., 3rd. Mechanistic inferences on metabolic dysfunction in posttraumatic stress disorder from an integrated model and multiomic analysis: role of glucocorticoid receptor sensitivity. Am. J. Physiol. Endocrinol. Metab. 2019;317:E879–E898. doi: 10.1152/ajpendo.00065.2019. - DOI - PMC - PubMed
-
- Yang R., Gautam A., Getnet D., Daigle B.J., Miller S., Misganaw B., Dean K.R., Kumar R., Muhie S., Wang K., et al. Epigenetic biotypes of post-traumatic stress disorder in war-zone exposed veteran and active duty males. Mol. Psychiatry. 2021;26:4300–4314. doi: 10.1038/s41380-020-00966-2. - DOI - PMC - PubMed
-
- Katrinli S., Stevens J., Wani A.H., Lori A., Kilaru V., van Rooij S.J.H., Hinrichs R., Powers A., Gillespie C.F., Michopoulos V., et al. Evaluating the impact of trauma and PTSD on epigenetic prediction of lifespan and neural integrity. Neuropsychopharmacology. 2020;45:1609–1616. doi: 10.1038/s41386-020-0700-5. - DOI - PMC - PubMed

