Hippocampo-cortical circuits for selective memory encoding, routing, and replay
- PMID: 37196658
- PMCID: PMC11146684
- DOI: 10.1016/j.neuron.2023.04.015
Hippocampo-cortical circuits for selective memory encoding, routing, and replay
Abstract
Traditionally considered a homogeneous cell type, hippocampal pyramidal cells have been recently shown to be highly diverse. However, how this cellular diversity relates to the different hippocampal network computations that support memory-guided behavior is not yet known. We show that the anatomical identity of pyramidal cells is a major organizing principle of CA1 assembly dynamics, the emergence of memory replay, and cortical projection patterns in rats. Segregated pyramidal cell subpopulations encoded trajectory and choice-specific information or tracked changes in reward configuration respectively, and their activity was selectively read out by different cortical targets. Furthermore, distinct hippocampo-cortical assemblies coordinated the reactivation of complementary memory representations. These findings reveal the existence of specialized hippocampo-cortical subcircuits and provide a cellular mechanism that supports the computational flexibility and memory capacities of these structures.
Keywords: cell assemblies; entorhinal cortex; hippocampus; memory reactivation; memory replay; oscillations; place cells; prefrontal cortex; sharp-wave ripples.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
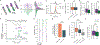
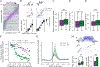




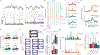
References
-
- O’Keefe J and Nadel L (1978). The Hippocampus as a Cognitive Map (Oxford: Clarendon Press; ).
-
- Wilson M and McNaughton B (1994). Reactivation of hippocampal ensemble memories during sleep. Science 265, 676–679. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

