EMC chaperone-CaV structure reveals an ion channel assembly intermediate
- PMID: 37196677
- PMCID: PMC10896479
- DOI: 10.1038/s41586-023-06175-5
EMC chaperone-CaV structure reveals an ion channel assembly intermediate
Abstract
Voltage-gated ion channels (VGICs) comprise multiple structural units, the assembly of which is required for function1,2. Structural understanding of how VGIC subunits assemble and whether chaperone proteins are required is lacking. High-voltage-activated calcium channels (CaVs)3,4 are paradigmatic multisubunit VGICs whose function and trafficking are powerfully shaped by interactions between pore-forming CaV1 or CaV2 CaVα1 (ref. 3), and the auxiliary CaVβ5 and CaVα2δ subunits6,7. Here we present cryo-electron microscopy structures of human brain and cardiac CaV1.2 bound with CaVβ3 to a chaperone-the endoplasmic reticulum membrane protein complex (EMC)8,9-and of the assembled CaV1.2-CaVβ3-CaVα2δ-1 channel. These structures provide a view of an EMC-client complex and define EMC sites-the transmembrane (TM) and cytoplasmic (Cyto) docks; interaction between these sites and the client channel causes partial extraction of a pore subunit and splays open the CaVα2δ-interaction site. The structures identify the CaVα2δ-binding site for gabapentinoid anti-pain and anti-anxiety drugs6, show that EMC and CaVα2δ interactions with the channel are mutually exclusive, and indicate that EMC-to-CaVα2δ hand-off involves a divalent ion-dependent step and CaV1.2 element ordering. Disruption of the EMC-CaV complex compromises CaV function, suggesting that the EMC functions as a channel holdase that facilitates channel assembly. Together, the structures reveal a CaV assembly intermediate and EMC client-binding sites that could have wide-ranging implications for the biogenesis of VGICs and other membrane proteins.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Figures

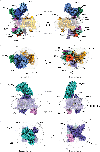



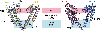








References
-
- Nanou E & Catterall WA Calcium channels, synaptic plasticity, and neuropsychiatric disease. Neuron 98, 466–481 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

