Multi-ancestry meta-analysis and fine-mapping in Alzheimer's disease
- PMID: 37198259
- PMCID: PMC10615750
- DOI: 10.1038/s41380-023-02089-w
Multi-ancestry meta-analysis and fine-mapping in Alzheimer's disease
Abstract
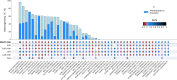
Genome-wide association studies (GWAS) of Alzheimer's disease are predominantly carried out in European ancestry individuals despite the known variation in genetic architecture and disease prevalence across global populations. We leveraged published GWAS summary statistics from European, East Asian, and African American populations, and an additional GWAS from a Caribbean Hispanic population using previously reported genotype data to perform the largest multi-ancestry GWAS meta-analysis of Alzheimer's disease and related dementias to date. This method allowed us to identify two independent novel disease-associated loci on chromosome 3. We also leveraged diverse haplotype structures to fine-map nine loci with a posterior probability >0.8 and globally assessed the heterogeneity of known risk factors across populations. Additionally, we compared the generalizability of multi-ancestry- and single-ancestry-derived polygenic risk scores in a three-way admixed Colombian population. Our findings highlight the importance of multi-ancestry representation in uncovering and understanding putative factors that contribute to risk of Alzheimer's disease and related dementias.
© 2023. The Author(s).
Conflict of interest statement
KL, DV, HL and MAN’s participation in this project was part of a competitive contract awarded to Data Tecnica International LLC by the National Institutes of Health to support open science research. MAN also currently serves on the scientific advisory board for Clover Therapeutics and is an advisor to Neuron23 Inc. PH is an employee at Alector and owns stock and options. JSY serves on the scientific advisory board for the Epstein Family Alzheimer’s Research Collaboration.
Figures