Targeting metabolism by B-raf inhibitors and diclofenac restrains the viability of BRAF-mutated thyroid carcinomas with Hif-1α-mediated glycolytic phenotype
- PMID: 37198319
- PMCID: PMC10338540
- DOI: 10.1038/s41416-023-02282-2
Targeting metabolism by B-raf inhibitors and diclofenac restrains the viability of BRAF-mutated thyroid carcinomas with Hif-1α-mediated glycolytic phenotype
Abstract
Background: B-raf inhibitors (BRAFi) are effective for BRAF-mutated papillary (PTC) and anaplastic (ATC) thyroid carcinomas, although acquired resistance impairs tumour cells' sensitivity and/or limits drug efficacy. Targeting metabolic vulnerabilities is emerging as powerful approach in cancer.
Methods: In silico analyses identified metabolic gene signatures and Hif-1α as glycolysis regulator in PTC. BRAF-mutated PTC, ATC and control thyroid cell lines were exposed to HIF1A siRNAs or chemical/drug treatments (CoCl2, EGF, HGF, BRAFi, MEKi and diclofenac). Genes/proteins expression, glucose uptake, lactate quantification and viability assays were used to investigate the metabolic vulnerability of BRAF-mutated cells.
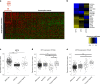
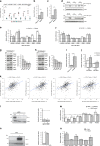
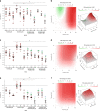
Results: A specific metabolic gene signature was identified as a hallmark of BRAF-mutated tumours, which display a glycolytic phenotype, characterised by enhanced glucose uptake, lactate efflux and increased expression of Hif-1α-modulated glycolytic genes. Indeed, Hif-1α stabilisation counteracts the inhibitory effects of BRAFi on these genes and on cell viability. Interestingly, targeting metabolic routes with BRAFi and diclofenac combination we could restrain the glycolytic phenotype and synergistically reduce tumour cells' viability.
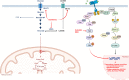
Conclusion: The identification of a metabolic vulnerability of BRAF-mutated carcinomas and the capacity BRAFi and diclofenac combination to target metabolism open new therapeutic perspectives in maximising drug efficacy and reducing the onset of secondary resistance and drug-related toxicity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Combined BRAF and PIM1 inhibitory therapy for papillary thyroid carcinoma based on BRAFV600E regulation of PIM1: Synergistic effect and metabolic mechanisms.Neoplasia. 2024 Jun;52:100996. doi: 10.1016/j.neo.2024.100996. Epub 2024 Apr 8. Neoplasia. 2024. PMID: 38593698 Free PMC article.
-
Fibronectin Contributes to a BRAF Inhibitor-driven Invasive Phenotype in Thyroid Cancer through EGR1, Which Can Be Blocked by Inhibition of ERK1/2.Mol Cancer Res. 2023 Sep 1;21(9):867-880. doi: 10.1158/1541-7786.MCR-22-1031. Mol Cancer Res. 2023. PMID: 37219859 Free PMC article.
-
Evidence that one subset of anaplastic thyroid carcinomas are derived from papillary carcinomas due to BRAF and p53 mutations.Cancer. 2005 Jun 1;103(11):2261-8. doi: 10.1002/cncr.21073. Cancer. 2005. PMID: 15880523 Review.
-
BRAF(V600E) mutation influences hypoxia-inducible factor-1alpha expression levels in papillary thyroid cancer.Mod Pathol. 2010 Aug;23(8):1052-60. doi: 10.1038/modpathol.2010.86. Epub 2010 May 14. Mod Pathol. 2010. PMID: 20473281
-
Efficacy and safety of BRAF/MEK inhibitors in BRAFV600E-mutated anaplastic thyroid cancer: a systematic review and meta-analysis.Endocrine. 2024 Oct;86(1):284-292. doi: 10.1007/s12020-024-03845-w. Epub 2024 May 6. Endocrine. 2024. PMID: 38709445
Cited by
-
Unraveling BRAF alterations: molecular insights to circumvent therapeutic resistance across cancer types.Cancer Drug Resist. 2025 Mar 24;8:14. doi: 10.20517/cdr.2024.213. eCollection 2025. Cancer Drug Resist. 2025. PMID: 40201310 Free PMC article.
-
Reprogramming of Thyroid Cancer Metabolism: from Mechanism to Therapeutic Strategy.Mol Cancer. 2025 Mar 11;24(1):74. doi: 10.1186/s12943-025-02263-4. Mol Cancer. 2025. PMID: 40069775 Free PMC article. Review.
-
Targeting Hypoxia-Inducible Factor-1 (HIF-1) in Cancer: Emerging Therapeutic Strategies and Pathway Regulation.Pharmaceuticals (Basel). 2024 Feb 1;17(2):195. doi: 10.3390/ph17020195. Pharmaceuticals (Basel). 2024. PMID: 38399410 Free PMC article. Review.
-
Revolutionizing Cancer Treatment: Unveiling New Frontiers by Targeting the (Un)Usual Suspects.Cancers (Basel). 2023 Dec 27;16(1):132. doi: 10.3390/cancers16010132. Cancers (Basel). 2023. PMID: 38201558 Free PMC article.
-
Toward Systems-Level Metabolic Analysis in Endocrine Disorders and Cancer.Endocrinol Metab (Seoul). 2023 Dec;38(6):619-630. doi: 10.3803/EnM.2023.1814. Epub 2023 Nov 21. Endocrinol Metab (Seoul). 2023. PMID: 37989266 Free PMC article. Review.
References
-
- Fendt SM, Frezza C, Erez A. Targeting metabolic plasticity and flexibility dynamics for cancer therapy. Cancer Discov. 2020;10:1797–807. doi: 10.1158/2159-8290.CD-20-0844. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

