Salvage of ribose from uridine or RNA supports glycolysis in nutrient-limited conditions
- PMID: 37198474
- PMCID: PMC10229423
- DOI: 10.1038/s42255-023-00774-2
Salvage of ribose from uridine or RNA supports glycolysis in nutrient-limited conditions
Abstract
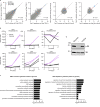
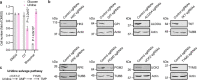
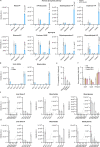
Glucose is vital for life, serving as both a source of energy and carbon building block for growth. When glucose is limiting, alternative nutrients must be harnessed. To identify mechanisms by which cells can tolerate complete loss of glucose, we performed nutrient-sensitized genome-wide genetic screens and a PRISM growth assay across 482 cancer cell lines. We report that catabolism of uridine from the medium enables the growth of cells in the complete absence of glucose. While previous studies have shown that uridine can be salvaged to support pyrimidine synthesis in the setting of mitochondrial oxidative phosphorylation deficiency1, our work demonstrates that the ribose moiety of uridine or RNA can be salvaged to fulfil energy requirements via a pathway based on: (1) the phosphorylytic cleavage of uridine by uridine phosphorylase UPP1/UPP2 into uracil and ribose-1-phosphate (R1P), (2) the conversion of uridine-derived R1P into fructose-6-P and glyceraldehyde-3-P by the non-oxidative branch of the pentose phosphate pathway and (3) their glycolytic utilization to fuel ATP production, biosynthesis and gluconeogenesis. Capacity for glycolysis from uridine-derived ribose appears widespread, and we confirm its activity in cancer lineages, primary macrophages and mice in vivo. An interesting property of this pathway is that R1P enters downstream of the initial, highly regulated steps of glucose transport and upper glycolysis. We anticipate that 'uridine bypass' of upper glycolysis could be important in the context of disease and even exploited for therapeutic purposes.
© 2023. The Author(s).
Conflict of interest statement
V.K.M. is a paid scientific advisor to 5AM Ventures. O.S.S. was a paid consultant for Proteinaceous Inc. D.E.F. has a financial interest in Soltego, a company developing salt-inducible kinase inhibitors for topical skin-darkening treatments that might be used for a broad set of human applications. The interests of D.E.F. were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict-of-interest policies. The remaining authors declare no competing interests.
Figures



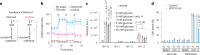





References
-
- Loffler M, Wenzel A, Schneider F. Cytokinetic studies on the switch from glucose to uridine metabolism, and vice versa, of Ehrlich ascites tumour cells in vitro. Cell Tissue Kinet. 1987;20:181–190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

