Ancient gene linkages support ctenophores as sister to other animals
- PMID: 37198475
- PMCID: PMC10232365
- DOI: 10.1038/s41586-023-05936-6
Ancient gene linkages support ctenophores as sister to other animals
Abstract
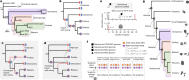
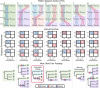
A central question in evolutionary biology is whether sponges or ctenophores (comb jellies) are the sister group to all other animals. These alternative phylogenetic hypotheses imply different scenarios for the evolution of complex neural systems and other animal-specific traits1-6. Conventional phylogenetic approaches based on morphological characters and increasingly extensive gene sequence collections have not been able to definitively answer this question7-11. Here we develop chromosome-scale gene linkage, also known as synteny, as a phylogenetic character for resolving this question12. We report new chromosome-scale genomes for a ctenophore and two marine sponges, and for three unicellular relatives of animals (a choanoflagellate, a filasterean amoeba and an ichthyosporean) that serve as outgroups for phylogenetic analysis. We find ancient syntenies that are conserved between animals and their close unicellular relatives. Ctenophores and unicellular eukaryotes share ancestral metazoan patterns, whereas sponges, bilaterians, and cnidarians share derived chromosomal rearrangements. Conserved syntenic characters unite sponges with bilaterians, cnidarians, and placozoans in a monophyletic clade to the exclusion of ctenophores, placing ctenophores as the sister group to all other animals. The patterns of synteny shared by sponges, bilaterians, and cnidarians are the result of rare and irreversible chromosome fusion-and-mixing events that provide robust and unambiguous phylogenetic support for the ctenophore-sister hypothesis. These findings provide a new framework for resolving deep, recalcitrant phylogenetic problems and have implications for our understanding of animal evolution.
© 2023. The Author(s).
Conflict of interest statement
R.E.G. and D.S.R. are paid consultants and equity holders of Dovetail Genomics. D.T.S. is a shareholder of Pacific Biosciences of California. The other authors declare no competing interests.
Figures