Uridine-derived ribose fuels glucose-restricted pancreatic cancer
- PMID: 37198494
- PMCID: PMC10232363
- DOI: 10.1038/s41586-023-06073-w
Uridine-derived ribose fuels glucose-restricted pancreatic cancer
Abstract
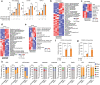
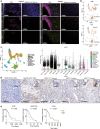
Pancreatic ductal adenocarcinoma (PDA) is a lethal disease notoriously resistant to therapy1,2. This is mediated in part by a complex tumour microenvironment3, low vascularity4, and metabolic aberrations5,6. Although altered metabolism drives tumour progression, the spectrum of metabolites used as nutrients by PDA remains largely unknown. Here we identified uridine as a fuel for PDA in glucose-deprived conditions by assessing how more than 175 metabolites impacted metabolic activity in 21 pancreatic cell lines under nutrient restriction. Uridine utilization strongly correlated with the expression of uridine phosphorylase 1 (UPP1), which we demonstrate liberates uridine-derived ribose to fuel central carbon metabolism and thereby support redox balance, survival and proliferation in glucose-restricted PDA cells. In PDA, UPP1 is regulated by KRAS-MAPK signalling and is augmented by nutrient restriction. Consistently, tumours expressed high UPP1 compared with non-tumoural tissues, and UPP1 expression correlated with poor survival in cohorts of patients with PDA. Uridine is available in the tumour microenvironment, and we demonstrated that uridine-derived ribose is actively catabolized in tumours. Finally, UPP1 deletion restricted the ability of PDA cells to use uridine and blunted tumour growth in immunocompetent mouse models. Our data identify uridine utilization as an important compensatory metabolic process in nutrient-deprived PDA cells, suggesting a novel metabolic axis for PDA therapy.
© 2023. The Author(s).
Conflict of interest statement
C.A.L. has received consulting fees from Astellas Pharmaceuticals, Odyssey Therapeutics, and T-Knife Therapeutics, and is an inventor on patents pertaining to KRAS-regulated metabolic pathways, redox control pathways in pancreatic cancer, and targeting the GOT1-pathway as a therapeutic approach (US patent 2015126580-A1, 05/07/2015; US patent 20190136238, 05/09/2019; international patent WO2013177426-A2, 04/23/2015). A.S. received grants from Merck, Pierre Fabre and Bristol Myers Squibb and is an inventor on patents on colorectal cancer classification with differential prognosis and personalized therapeutic responses (PCT/IB2013/060416); prognostic and treatment response prediction in gastric cancer (priority patent CSC/BP7295892); patient classification and prognostic method (GEP-NET) (priority patent EP18425009.0); and molecular predictors of therapeutic response to specific anti-cancer agents (US9506926B2).
Figures












Comment in
-
Uridine: as sweet as sugar for some cells?Cell Res. 2023 Dec;33(12):898-899. doi: 10.1038/s41422-023-00860-w. Cell Res. 2023. PMID: 37567975 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- K99 CA267176/CA/NCI NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- P30 CA014599/CA/NCI NIH HHS/United States
- P30 DK056341/DK/NIDDK NIH HHS/United States
- T32 CA009594/CA/NCI NIH HHS/United States
- R01 CA248160/CA/NCI NIH HHS/United States
- K08 CA234222/CA/NCI NIH HHS/United States
- T32 GM139774/GM/NIGMS NIH HHS/United States
- R37 CA262209/CA/NCI NIH HHS/United States
- IK2 BX005875/BX/BLRD VA/United States
- R37 CA237421/CA/NCI NIH HHS/United States
- T32 CA140044/CA/NCI NIH HHS/United States
- R01 CA198074/CA/NCI NIH HHS/United States
- R00 CA241357/CA/NCI NIH HHS/United States
- T32 HD007505/HD/NICHD NIH HHS/United States
- R25 GM143298/GM/NIGMS NIH HHS/United States
- R01 CA151588/CA/NCI NIH HHS/United States
- R25 GM086262/GM/NIGMS NIH HHS/United States
- R01 CA244931/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

