Ex vivo factor VIII-modified proliferating human hepatocytes therapy for haemophilia A
- PMID: 37199059
- PMCID: PMC10212703
- DOI: 10.1111/cpr.13467
Ex vivo factor VIII-modified proliferating human hepatocytes therapy for haemophilia A
Abstract
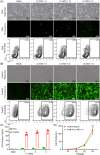
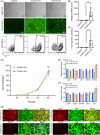
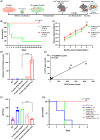
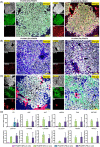
Ex vivo gene manipulation in human hepatocytes is a promising therapeutic strategy in the treatment of inherited liver diseases. However, a major limitation is the lack of a highly efficient and safe genetic manipulation system for transplantable primary human hepatocytes (PHHs). Here, we reported that proliferating human hepatocytes (ProliHHs) cultured in vitro showed high susceptibility to lentivirus-mediated genetic modification and maintained cellular phenotypes after lentiviral infection. Human factor VIII expression was introduced through F8-Lentivirus-mediated transduction of ProliHHs followed by xenotransplantation into immunocompromised haemophilia A mice. We demonstrated that these F8-modified ProliHHs could effectively repopulate the mouse liver, resulting in therapeutic benefits in mouse models. Furthermore, no genotoxicity was detected in F8-modified ProliHHs using lentiviral integration site analysis. Thus, this study demonstrated, for the first time, the feasibility and safety of lentiviral modification in ProliHHs to induce the expression of coagulation factor VIII in the treatment of haemophilia A.
© 2023 The Authors. Cell Proliferation published by Beijing Institute for Stem Cell and Regenerative Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Transduction of modified factor VIII gene improves lentiviral gene therapy efficacy for hemophilia A.J Biol Chem. 2021 Dec;297(6):101397. doi: 10.1016/j.jbc.2021.101397. Epub 2021 Nov 10. J Biol Chem. 2021. PMID: 34774524 Free PMC article.
-
Preclinical Development of a Hematopoietic Stem and Progenitor Cell Bioengineered Factor VIII Lentiviral Vector Gene Therapy for Hemophilia A.Hum Gene Ther. 2018 Oct;29(10):1183-1201. doi: 10.1089/hum.2018.137. Hum Gene Ther. 2018. PMID: 30160169 Free PMC article.
-
Functional Proliferating Human Hepatocytes: In Vitro Hepatocyte Model for Drug Metabolism, Excretion, and Toxicity.Drug Metab Dispos. 2021 Apr;49(4):305-313. doi: 10.1124/dmd.120.000275. Epub 2021 Feb 1. Drug Metab Dispos. 2021. PMID: 33526515
-
Haemophilia A gene therapy.Haemophilia. 1998 Jul;4(4):380-8. doi: 10.1046/j.1365-2516.1998.440380.x. Haemophilia. 1998. PMID: 9873758 Review.
-
Haemophilia gene therapy: From trailblazer to gamechanger.Haemophilia. 2018 May;24 Suppl 6:50-59. doi: 10.1111/hae.13494. Haemophilia. 2018. PMID: 29878653 Review.
Cited by
-
Expansion of human hepatocytes and their application in three-dimensional culture and genetic manipulation.Nat Protoc. 2025 Jul 25. doi: 10.1038/s41596-025-01211-2. Online ahead of print. Nat Protoc. 2025. PMID: 40715744 Review.
-
Silicone cryogel skeletons enhance the survival and mechanical integrity of hydrogel-encapsulated cell therapies.Sci Adv. 2024 Apr 5;10(14):eadk5949. doi: 10.1126/sciadv.adk5949. Epub 2024 Apr 5. Sci Adv. 2024. PMID: 38578991 Free PMC article.
-
Comparing acute versus AIDS ART initiation on HIV-1 integration sites and clonal expansion.Signal Transduct Target Ther. 2025 Jan 10;10(1):23. doi: 10.1038/s41392-024-02113-7. Signal Transduct Target Ther. 2025. PMID: 39788938 Free PMC article.
References
-
- Peters R, Harris T. Advances and innovations in haemophilia treatment. Nat Rev Drug Discov. 2018;17(7):493‐508. - PubMed
-
- Berntorp E, Shapiro AD. Modern haemophilia care. Lancet. 2012;379(9824):1447‐1456. - PubMed
-
- Blanchette VS, Key NS, Ljung LR, et al. Definitions in hemophilia: communication from the SSC of the ISTH. J Thromb Haemost. 2014;12(11):1935‐1939. - PubMed
-
- Berntorp E, Fischer K, Hart DP, et al. Haemophilia. Nat Rev Dis Primers. 2021;7(1):45. - PubMed
-
- Knight R, Stanley S, Wong M, Dolan G. Hemophilia therapy and blood‐borne pathogen risk. Semin Thromb Hemost. 2006;32(Suppl 2):3‐9. - PubMed
MeSH terms
Substances
Grants and funding
- E019710101/Special Research Assistant Project of the Chinese Academy of Sciences
- XDA16020201/"Strategic Priority Research Program" of the Chinese Academy of Sciences
- 2019M660101/China Postdoctoral Science Foundation
- 2019TQ0339/China Postdoctoral Science Foundation
- 32001070/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

