Engineered human Diamond-Blackfan anemia disease model confirms therapeutic effects of clinically applicable lentiviral vector at single-cell resolution
- PMID: 37199130
- PMCID: PMC10620578
- DOI: 10.3324/haematol.2022.282068
Engineered human Diamond-Blackfan anemia disease model confirms therapeutic effects of clinically applicable lentiviral vector at single-cell resolution
Abstract
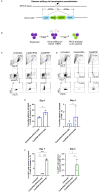
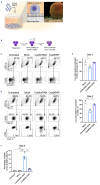
Diamond-Blackfan anemia is a rare genetic bone marrow failure disorder which is usually caused by mutations in ribosomal protein genes. In the present study, we generated a traceable RPS19-deficient cell model using CRISPR-Cas9 and homology-directed repair to investigate the therapeutic effects of a clinically applicable lentiviral vector at single-cell resolution. We developed a gentle nanostraw delivery platform to edit the RPS19 gene in primary human cord bloodderived CD34+ hematopoietic stem and progenitor cells. The edited cells showed expected impaired erythroid differentiation phenotype, and a specific erythroid progenitor with abnormal cell cycle status accompanied by enrichment of TNFα/NF-κB and p53 signaling pathways was identified by single-cell RNA sequencing analysis. The therapeutic vector could rescue the abnormal erythropoiesis by activating cell cycle-related signaling pathways and promoted red blood cell production. Overall, these results establish nanostraws as a gentle option for CRISPR-Cas9- based gene editing in sensitive primary hematopoietic stem and progenitor cells, and provide support for future clinical investigations of the lentiviral gene therapy strategy.
Figures




Comment in
-
Gene therapy for congenital marrow failure syndromes - no longer grasping at straws?Haematologica. 2023 Nov 1;108(11):2880-2882. doi: 10.3324/haematol.2023.283462. Haematologica. 2023. PMID: 37317900 Free PMC article. No abstract available.
References
-
- Iskander D, Wang G, Heuston EF, et al. . Single-cell profiling of human bone marrow progenitors reveals mechanisms of failing erythropoiesis in Diamond-Blackfan anemia. Sci Transl Med. 2021;13(610):eabf0113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

