Two Permeases Associated with the Multifunctional CtaP Cysteine Transport System in Listeria monocytogenes Play Distinct Roles in Pathogenesis
- PMID: 37199604
- PMCID: PMC10269559
- DOI: 10.1128/spectrum.03317-22
Two Permeases Associated with the Multifunctional CtaP Cysteine Transport System in Listeria monocytogenes Play Distinct Roles in Pathogenesis
Abstract
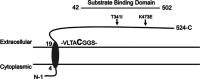
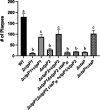
The soil-dwelling bacterium Listeria monocytogenes survives a multitude of conditions when residing in the outside environment and as a pathogen within host cells. Key to survival within the infected mammalian host is the expression of bacterial gene products necessary for nutrient acquisition. Similar to many bacteria, L. monocytogenes uses peptide import to acquire amino acids. Peptide transport systems play an important role in nutrient uptake as well as in additional functions that include bacterial quorum sensing and signal transduction, recycling of peptidoglycan fragments, adherence to eukaryotic cells, and alterations in antibiotic susceptibility. It has been previously described that CtaP, encoded by lmo0135, is a multifunctional protein associated with activities that include cysteine transport, resistance to acid, membrane integrity, and bacterial adherence to host cells. ctaP is located next to two genes predicted to encode membrane-bound permeases lmo0136 and lmo0137, termed CtpP1 and CtpP2, respectively. Here, we show that CtpP1 and CtpP2 are required for bacterial growth in the presence of low concentrations of cysteine and for virulence in mouse infection models. Taken together, the data identify distinct nonoverlapping roles for two related permeases that are important for the growth and survival of L. monocytogenes within host cells. IMPORTANCE Bacterial peptide transport systems are important for nutrient uptake and may additionally function in a variety of other roles, including bacterial communication, signal transduction, and bacterial adherence to eukaryotic cells. Peptide transport systems often consist of a substrate-binding protein associated with a membrane-spanning permease. The environmental bacterial pathogen Listeria monocytogenes uses the substrate-binding protein CtaP not only for cysteine transport but also for resistance to acid, maintenance of membrane integrity, and bacterial adherence to host cells. In this study, we demonstrate complementary yet distinct functional roles for two membrane permeases, CtpP1 and CtpP2, that are encoded by genes linked to ctaP and that contribute to bacterial growth, invasion, and pathogenicity.
Keywords: ABC transporters; CtaP; PrfA; bacterial adhesion; cysteine transport; virulence; virulence factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

