Burkholderia contaminans Bacteriophage CSP3 Requires O-Antigen Polysaccharides for Infection
- PMID: 37199610
- PMCID: PMC10269572
- DOI: 10.1128/spectrum.05332-22
Burkholderia contaminans Bacteriophage CSP3 Requires O-Antigen Polysaccharides for Infection
Abstract
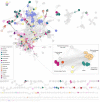
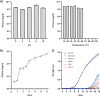
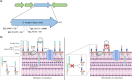
The Burkholderia cepacia complex is a group of opportunistic pathogens that cause both severe acute and chronic respiratory infections. Due to their large genomes containing multiple intrinsic and acquired antimicrobial resistance mechanisms, treatment is often difficult and prolonged. One alternative to traditional antibiotics for treatment of bacterial infections is bacteriophages. Therefore, the characterization of bacteriophages infective for the Burkholderia cepacia complex is critical to determine their suitability for any future use. Here, we describe the isolation and characterization of novel phage, CSP3, infective against a clinical isolate of Burkholderia contaminans. CSP3 is a new member of the Lessievirus genus that targets various Burkholderia cepacia complex organisms. Single nucleotide polymorphism (SNP) analysis of CSP3-resistant B. contaminans showed that mutations to the O-antigen ligase gene, waaL, consequently inhibited CSP3 infection. This mutant phenotype is predicted to result in the loss of cell surface O-antigen, contrary to a related phage that requires the inner core of the lipopolysaccharide for infection. Additionally, liquid infection assays showed that CSP3 provides suppression of B. contaminans growth for up to 14 h. Despite the inclusion of genes that are typical of the phage lysogenic life cycle, we saw no evidence of CSP3's ability to lysogenize. Continuation of phage isolation and characterization is crucial in developing large and diverse phage banks for global usage in cases of antibiotic-resistant bacterial infections. IMPORTANCE Amid the global antibiotic resistance crisis, novel antimicrobials are needed to treat problematic bacterial infections, including those from the Burkholderia cepacia complex. One such alternative is the use of bacteriophages; however, a lot is still unknown about their biology. Bacteriophage characterization studies are of high importance for building phage banks, as future work in developing treatments such as phage cocktails should require well-characterized phages. Here, we report the isolation and characterization of a novel Burkholderia contaminans phage that requires the O-antigen for infection, a distinct phenotype seen among other related phages. Our findings presented in this article expand on the ever-evolving phage biology field, uncovering unique phage-host relationships and mechanisms of infection.
Keywords: Burkholderia cepacia complex; O-antigen; bacteriophage; characterization; phage; phage characterization; phage therapy; receptor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Isolation of a PRD1-like phage uncovers the carriage of three putative conjugative plasmids in clinical Burkholderia contaminans.Res Microbiol. 2024 Jul-Aug;175(5-6):104202. doi: 10.1016/j.resmic.2024.104202. Epub 2024 Apr 4. Res Microbiol. 2024. PMID: 38582389
-
Synergistic Interactions among Burkholderia cepacia Complex-Targeting Phages Reveal a Novel Therapeutic Role for Lysogenization-Capable Phages.Microbiol Spectr. 2023 Jun 15;11(3):e0443022. doi: 10.1128/spectrum.04430-22. Epub 2023 May 17. Microbiol Spectr. 2023. PMID: 37195168 Free PMC article.
-
Harnessing the Diversity of Burkholderia spp. Prophages for Therapeutic Potential.Cells. 2024 Feb 29;13(5):428. doi: 10.3390/cells13050428. Cells. 2024. PMID: 38474392 Free PMC article.
-
The promise of bacteriophage therapy for Burkholderia cepacia complex respiratory infections.Front Cell Infect Microbiol. 2012 Jan 20;1:27. doi: 10.3389/fcimb.2011.00027. eCollection 2011. Front Cell Infect Microbiol. 2012. PMID: 22919592 Free PMC article. Review.
-
Advances in Phage Therapy: Targeting the Burkholderia cepacia Complex.Viruses. 2021 Jul 9;13(7):1331. doi: 10.3390/v13071331. Viruses. 2021. PMID: 34372537 Free PMC article. Review.
Cited by
-
A flagella-dependent Burkholderia jumbo phage controls rice seedling rot and steers Burkholderia glumae toward reduced virulence in rice seedlings.mBio. 2025 Mar 12;16(3):e0281424. doi: 10.1128/mbio.02814-24. Epub 2025 Jan 27. mBio. 2025. PMID: 39868782 Free PMC article.
-
Bacteriophage therapy to combat MDR non-fermenting Gram-negative bacteria causing nosocomial infections: recent progress and challenges.Naunyn Schmiedebergs Arch Pharmacol. 2025 Jun 6. doi: 10.1007/s00210-025-04345-y. Online ahead of print. Naunyn Schmiedebergs Arch Pharmacol. 2025. PMID: 40478338 Review.
-
Phage therapy to treat cystic fibrosis Burkholderia cepacia complex lung infections: perspectives and challenges.Front Microbiol. 2024 Oct 18;15:1476041. doi: 10.3389/fmicb.2024.1476041. eCollection 2024. Front Microbiol. 2024. PMID: 39493847 Free PMC article. Review.
-
Characterization and Abundance of Plasmid-Dependent Alphatectivirus Bacteriophages.Microb Ecol. 2024 Jun 27;87(1):85. doi: 10.1007/s00248-024-02401-3. Microb Ecol. 2024. PMID: 38935220 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

