Human Anelloviruses: Influence of Demographic Factors, Recombination, and Worldwide Diversity
- PMID: 37199659
- PMCID: PMC10269794
- DOI: 10.1128/spectrum.04928-22
Human Anelloviruses: Influence of Demographic Factors, Recombination, and Worldwide Diversity
Abstract
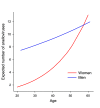
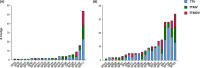
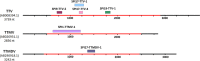
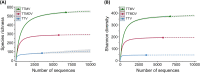
Anelloviruses represent the major and most diverse component of the healthy human virome, referred to as the anellome. In this study, we determined the anellome of 50 blood donors, forming two sex- and age-matched groups. Anelloviruses were detected in 86% of the donors. The number of detected anelloviruses increased with age and was approximately twice as high in men as in women. A total of 349 complete or nearly complete genomes were classified as belonging to torque teno virus (TTV), torque teno mini virus (TTMV), and torque teno midi virus (TTMDV) anellovirus genera (197, 88, and 64 sequences, respectively). Most donors had intergenus (69.8%) or intragenus (72.1%) coinfections. Despite the limited number of sequences, intradonor recombination analysis showed 6 intragenus recombination events in ORF1. As thousands of anellovirus sequences have been described recently, we finally analyzed the global diversity of human anelloviruses. Species richness and diversity were close to saturation in each anellovirus genus. Recombination was found to be the main factor promoting diversity, although its effect was significantly lower in TTV than in TTMV and TTMDV. Overall, our results suggest that differences in diversity between genera may be caused by variations in the relative contribution of recombination. IMPORTANCE Anelloviruses are the most common human infectious viruses and are considered essentially harmless. Compared to other human viruses, they are characterized by enormous diversity, and recombination is suggested to play an important role in their diversification and evolution. Here, by analyzing the composition of the plasma anellome of 50 blood donors, we find that recombination is also a determinant of viral evolution at the intradonor level. On a larger scale, analysis of anellovirus sequences currently available in databases shows that their diversity is close to saturation and differs among the three human anellovirus genera and that recombination is the main factor explaining this intergenus variability. Global characterization of anellovirus diversity could provide clues about possible associations between certain virus variants and pathologies, as well as facilitate the implementation of unbiased PCR-based detection protocols, which may be relevant for using anelloviruses as endogenous markers of immune status.
Keywords: anellovirus; blood anellome; metagenomics; recombination; virome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

